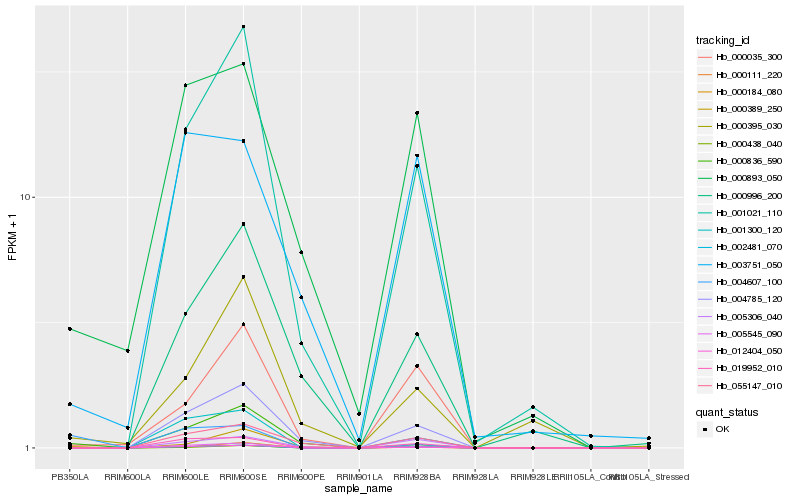
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_220 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000836_590 |
0.1879688875 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 3 |
Hb_000893_050 |
0.2073159737 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_004785_120 |
0.2120835763 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 5 |
Hb_001021_110 |
0.2255896543 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 6 |
Hb_055147_010 |
0.2316841185 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 7 |
Hb_000438_040 |
0.2418954046 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_000184_080 |
0.2423912492 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 9 |
Hb_004607_100 |
0.2426621284 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 10 |
Hb_005545_090 |
0.243634984 |
- |
- |
- |
| 11 |
Hb_000996_200 |
0.252153211 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 12 |
Hb_002481_070 |
0.2523766489 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 13 |
Hb_000389_250 |
0.2535700383 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 14 |
Hb_001300_120 |
0.2559303958 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |
| 15 |
Hb_000395_030 |
0.2569055689 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 16 |
Hb_003751_050 |
0.2572905368 |
- |
- |
PREDICTED: amino acid permease 4-like [Jatropha curcas] |
| 17 |
Hb_005306_040 |
0.2574304296 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 18 |
Hb_000035_300 |
0.2580911264 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_019952_010 |
0.258547234 |
- |
- |
- |
| 20 |
Hb_012404_050 |
0.2601678893 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |