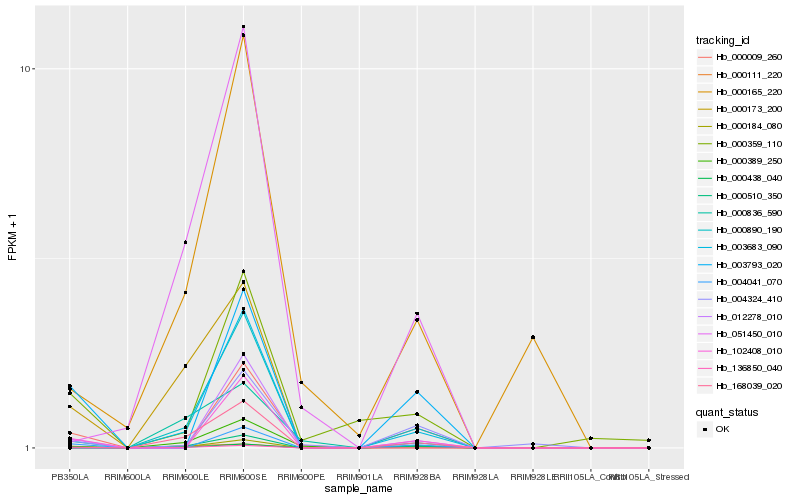
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_080 |
0.0 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 2 |
Hb_000438_040 |
0.1931114001 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_004041_070 |
0.2283551844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_136850_040 |
0.2361091141 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000389_250 |
0.2366076394 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 6 |
Hb_000111_220 |
0.2423912492 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003793_020 |
0.2435023251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_102408_010 |
0.2486640809 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 9 |
Hb_000009_260 |
0.2536844165 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000359_110 |
0.2640508257 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 42 [Theobroma cacao] |
| 11 |
Hb_000173_200 |
0.2666212526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000510_350 |
0.2666672892 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 13 |
Hb_004324_410 |
0.2681491429 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 14 |
Hb_000890_190 |
0.2733441504 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 15 |
Hb_000836_590 |
0.2741827191 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 16 |
Hb_012278_010 |
0.2745200491 |
- |
- |
- |
| 17 |
Hb_168039_020 |
0.2756699627 |
- |
- |
- |
| 18 |
Hb_051450_010 |
0.2812119386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003683_090 |
0.2843683756 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_000165_220 |
0.2851482378 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |