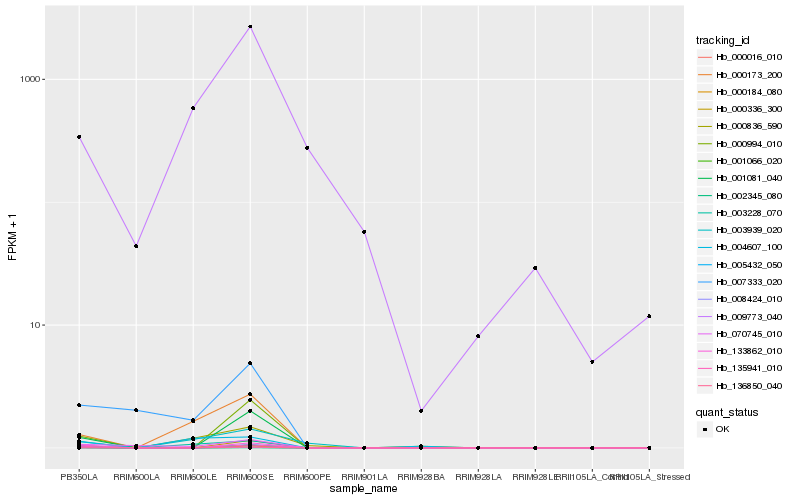
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136850_040 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_003228_070 |
0.1342880093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000173_200 |
0.1361481791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000184_080 |
0.2361091141 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 5 |
Hb_003939_020 |
0.2602783702 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 6 |
Hb_009773_040 |
0.2651884082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000336_300 |
0.2792591389 |
- |
- |
- |
| 8 |
Hb_000016_010 |
0.2813459821 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_008424_010 |
0.2815153791 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_001081_040 |
0.2841994139 |
- |
- |
hypothetical protein MIMGU_mgv11b021166mg, partial [Erythranthe guttata] |
| 11 |
Hb_007333_020 |
0.2843815752 |
- |
- |
- |
| 12 |
Hb_004607_100 |
0.2846852698 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 13 |
Hb_070745_010 |
0.2847967811 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 14 |
Hb_005432_050 |
0.2877866381 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 15 |
Hb_002345_080 |
0.2900428045 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_000994_010 |
0.2936383114 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 17 |
Hb_001066_020 |
0.2938898086 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_133862_010 |
0.2990057281 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 19 |
Hb_135941_010 |
0.3001764775 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 20 |
Hb_000836_590 |
0.300746851 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |