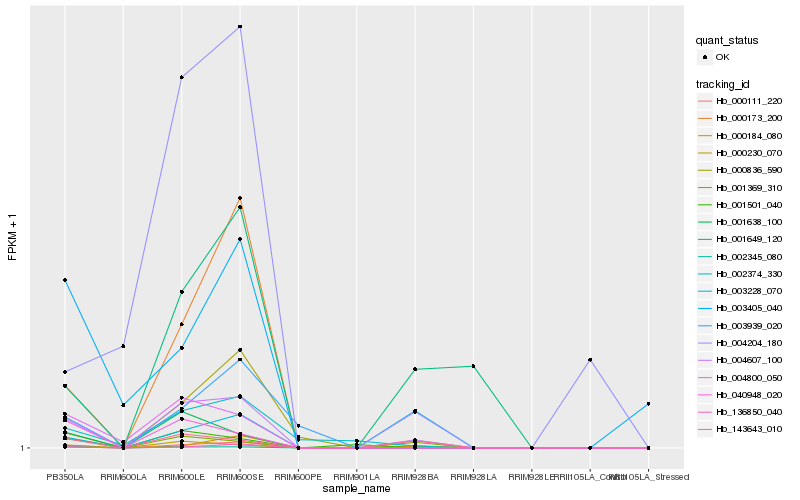
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 2 |
Hb_000230_070 |
0.2061298915 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 3 |
Hb_003228_070 |
0.2065271211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_143643_010 |
0.2174420648 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC103967596 [Pyrus x bretschneideri] |
| 5 |
Hb_002345_080 |
0.2222151264 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000111_220 |
0.2426621284 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_004800_050 |
0.2537694688 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 8 |
Hb_001369_310 |
0.2621659321 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 9 |
Hb_001638_100 |
0.2651413593 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 [Vitis vinifera] |
| 10 |
Hb_000173_200 |
0.2662527088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_040948_020 |
0.2701757161 |
- |
- |
- |
| 12 |
Hb_136850_040 |
0.2846852698 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_002374_330 |
0.2853387443 |
- |
- |
- |
| 14 |
Hb_000184_080 |
0.2958931362 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 15 |
Hb_001501_040 |
0.2975787766 |
- |
- |
PREDICTED: uncharacterized protein LOC102663792 [Glycine max] |
| 16 |
Hb_000836_590 |
0.298038955 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 17 |
Hb_003405_040 |
0.2999683541 |
- |
- |
PREDICTED: uncharacterized protein LOC105121978 [Populus euphratica] |
| 18 |
Hb_001649_120 |
0.3017193857 |
- |
- |
- |
| 19 |
Hb_003939_020 |
0.3245925856 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 20 |
Hb_004204_180 |
0.3312622344 |
- |
- |
- |