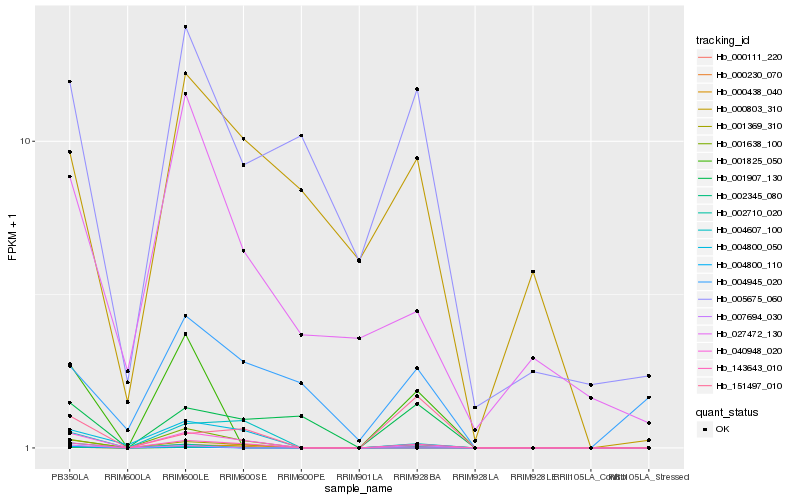
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143643_010 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC103967596 [Pyrus x bretschneideri] |
| 2 |
Hb_000230_070 |
0.0766686222 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 3 |
Hb_001369_310 |
0.1340572702 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 4 |
Hb_004607_100 |
0.2174420648 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 5 |
Hb_000111_220 |
0.3061580488 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_151497_010 |
0.3212587892 |
- |
- |
- |
| 7 |
Hb_001907_130 |
0.3215844445 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 8 |
Hb_040948_020 |
0.3218771376 |
- |
- |
- |
| 9 |
Hb_001638_100 |
0.3246007271 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 [Vitis vinifera] |
| 10 |
Hb_001825_050 |
0.3351926019 |
- |
- |
- |
| 11 |
Hb_002345_080 |
0.3358968424 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_004800_050 |
0.342156309 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 13 |
Hb_027472_130 |
0.3483152597 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 14 |
Hb_005675_060 |
0.3503805214 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 15 |
Hb_007694_030 |
0.353335378 |
- |
- |
hypothetical protein SORBIDRAFT_04g037485 [Sorghum bicolor] |
| 16 |
Hb_004945_020 |
0.358355362 |
- |
- |
PREDICTED: probable calcium-binding protein CML30 [Vitis vinifera] |
| 17 |
Hb_000803_310 |
0.3583617675 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002710_020 |
0.359384114 |
- |
- |
- |
| 19 |
Hb_004800_110 |
0.3595057752 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK4 [Jatropha curcas] |
| 20 |
Hb_000438_040 |
0.3654644515 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |