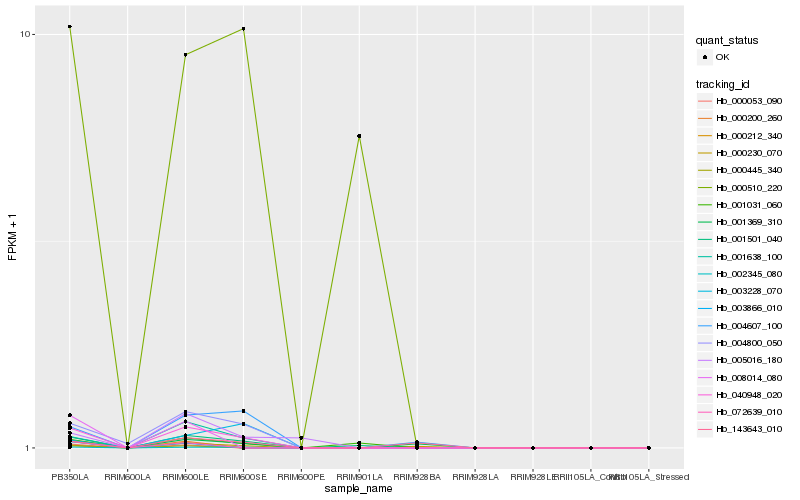
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_040948_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002345_080 |
0.1308463463 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001638_100 |
0.1506590675 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 [Vitis vinifera] |
| 4 |
Hb_004800_050 |
0.183153005 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 5 |
Hb_001501_040 |
0.237988771 |
- |
- |
PREDICTED: uncharacterized protein LOC102663792 [Glycine max] |
| 6 |
Hb_004607_100 |
0.2701757161 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 7 |
Hb_001369_310 |
0.2807009534 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 8 |
Hb_001031_060 |
0.3057934081 |
- |
- |
PREDICTED: uncharacterized protein LOC103940329 [Pyrus x bretschneideri] |
| 9 |
Hb_005016_180 |
0.3068616851 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 10 |
Hb_000230_070 |
0.3078206912 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 11 |
Hb_000510_220 |
0.3173058821 |
- |
- |
- |
| 12 |
Hb_143643_010 |
0.3218771376 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC103967596 [Pyrus x bretschneideri] |
| 13 |
Hb_003228_070 |
0.3279925319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000200_260 |
0.3282495693 |
- |
- |
PREDICTED: uncharacterized protein LOC104232531 isoform X2 [Nicotiana sylvestris] |
| 15 |
Hb_000212_340 |
0.3288331757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008014_080 |
0.3290046434 |
- |
- |
PREDICTED: uncharacterized protein LOC103848614 [Brassica rapa] |
| 17 |
Hb_072639_010 |
0.3291799096 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 18 |
Hb_000053_090 |
0.3292077339 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Populus euphratica] |
| 19 |
Hb_000445_340 |
0.32984888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003866_010 |
0.3314562665 |
- |
- |
Polyprotein, putative [Solanum demissum] |