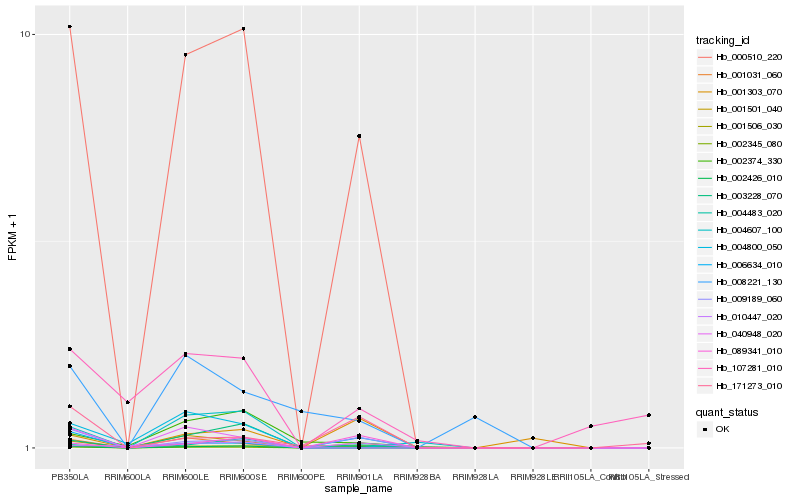
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_002426_010 |
0.0958781342 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 3 |
Hb_001031_060 |
0.1574250445 |
- |
- |
PREDICTED: uncharacterized protein LOC103940329 [Pyrus x bretschneideri] |
| 4 |
Hb_001501_040 |
0.1749407589 |
- |
- |
PREDICTED: uncharacterized protein LOC102663792 [Glycine max] |
| 5 |
Hb_009189_060 |
0.1942290309 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004483_020 |
0.2030832977 |
- |
- |
- |
| 7 |
Hb_006634_010 |
0.254707672 |
- |
- |
- |
| 8 |
Hb_089341_010 |
0.2776244924 |
- |
- |
Uncharacterized protein TCM_018339 [Theobroma cacao] |
| 9 |
Hb_002345_080 |
0.2826407552 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_002374_330 |
0.3024873292 |
- |
- |
- |
| 11 |
Hb_040948_020 |
0.3173058821 |
- |
- |
- |
| 12 |
Hb_004800_050 |
0.335974949 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 13 |
Hb_001506_030 |
0.3362338917 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_004607_100 |
0.3419910803 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 15 |
Hb_003228_070 |
0.3420766176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010447_020 |
0.344971914 |
- |
- |
PREDICTED: uncharacterized protein LOC105797464 [Gossypium raimondii] |
| 17 |
Hb_008221_130 |
0.3455134966 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 18 |
Hb_001303_070 |
0.3484690502 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X2 [Prunus mume] |
| 19 |
Hb_171273_010 |
0.3522642792 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 20 |
Hb_107281_010 |
0.3527469269 |
- |
- |
protein with unknown function [Ricinus communis] |