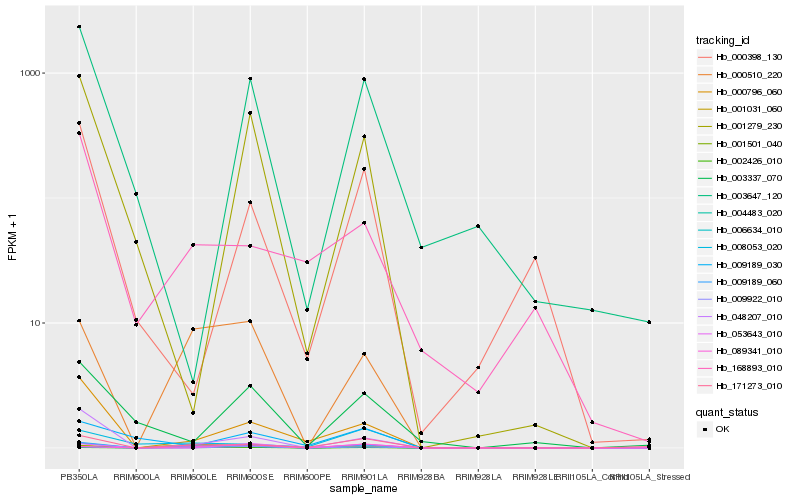
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009189_060 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006634_010 |
0.0665984824 |
- |
- |
- |
| 3 |
Hb_002426_010 |
0.1837806825 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_000510_220 |
0.1942290309 |
- |
- |
- |
| 5 |
Hb_171273_010 |
0.2156369704 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 6 |
Hb_001031_060 |
0.2203374859 |
- |
- |
PREDICTED: uncharacterized protein LOC103940329 [Pyrus x bretschneideri] |
| 7 |
Hb_048207_010 |
0.2323705475 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 8 |
Hb_000796_060 |
0.2597372623 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 9 |
Hb_089341_010 |
0.2856789441 |
- |
- |
Uncharacterized protein TCM_018339 [Theobroma cacao] |
| 10 |
Hb_008053_020 |
0.2921618462 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001279_230 |
0.3104540876 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 12 |
Hb_053643_010 |
0.3157300986 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 13 |
Hb_004483_020 |
0.3200955088 |
- |
- |
- |
| 14 |
Hb_003647_120 |
0.3211984575 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 15 |
Hb_003337_070 |
0.3213358041 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 16 |
Hb_168893_010 |
0.3238544442 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 17 |
Hb_000398_130 |
0.3290022465 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 18 |
Hb_001501_040 |
0.3300199435 |
- |
- |
PREDICTED: uncharacterized protein LOC102663792 [Glycine max] |
| 19 |
Hb_009189_030 |
0.3334416727 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 20 |
Hb_009922_010 |
0.3364006233 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |