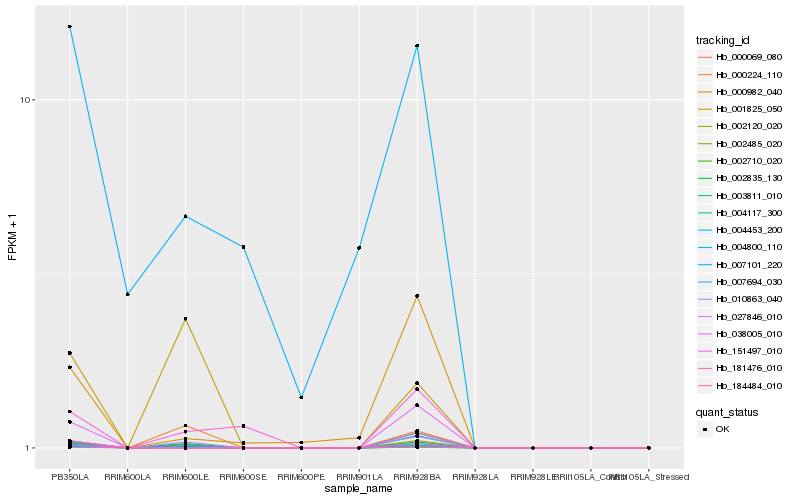
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 2 |
Hb_010863_040 |
0.0131232603 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 3 |
Hb_000069_080 |
0.0169798432 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 4 |
Hb_007694_030 |
0.1421179958 |
- |
- |
hypothetical protein SORBIDRAFT_04g037485 [Sorghum bicolor] |
| 5 |
Hb_002710_020 |
0.142851321 |
- |
- |
- |
| 6 |
Hb_004800_110 |
0.1487512221 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK4 [Jatropha curcas] |
| 7 |
Hb_000224_110 |
0.2225573794 |
- |
- |
- |
| 8 |
Hb_003811_010 |
0.2522758193 |
- |
- |
PREDICTED: uncharacterized protein LOC102611294 [Citrus sinensis] |
| 9 |
Hb_002485_020 |
0.2610899379 |
- |
- |
metal transport family protein [Populus trichocarpa] |
| 10 |
Hb_001825_050 |
0.2614258698 |
- |
- |
- |
| 11 |
Hb_151497_010 |
0.3102080672 |
- |
- |
- |
| 12 |
Hb_004453_200 |
0.3161668693 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 13 |
Hb_000982_040 |
0.3431940437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007101_220 |
0.3676184848 |
- |
- |
Gibberellin-regulated protein 3 precursor, putative [Ricinus communis] |
| 15 |
Hb_002120_020 |
0.3707881825 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 16 |
Hb_184484_010 |
0.3708342414 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_027846_010 |
0.3710683543 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 18 |
Hb_038005_010 |
0.3713382108 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 19 |
Hb_181476_010 |
0.3769427334 |
- |
- |
- |
| 20 |
Hb_004117_300 |
0.3789744309 |
- |
- |
- |