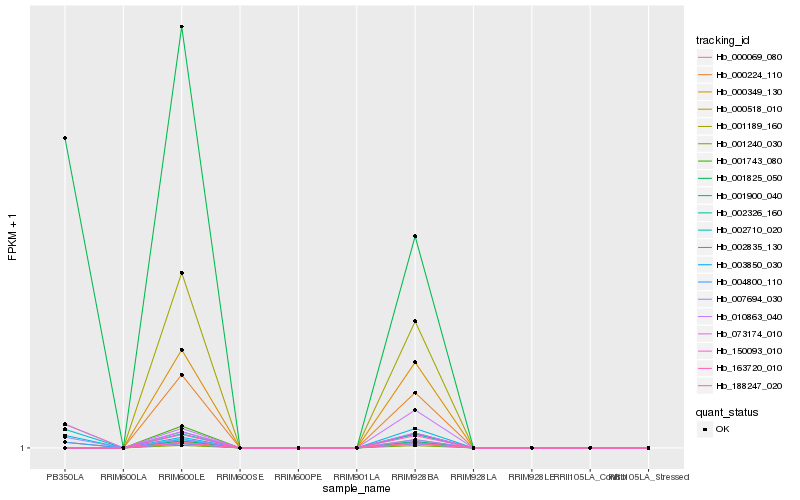
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000224_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_001825_050 |
0.2015741396 |
- |
- |
- |
| 3 |
Hb_002835_130 |
0.2225573794 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 4 |
Hb_007694_030 |
0.2290210222 |
- |
- |
hypothetical protein SORBIDRAFT_04g037485 [Sorghum bicolor] |
| 5 |
Hb_010863_040 |
0.2316135893 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 6 |
Hb_000069_080 |
0.2322587596 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 7 |
Hb_002710_020 |
0.2474970017 |
- |
- |
- |
| 8 |
Hb_004800_110 |
0.2519811602 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK4 [Jatropha curcas] |
| 9 |
Hb_001189_160 |
0.2818775163 |
- |
- |
- |
| 10 |
Hb_001743_080 |
0.2828767817 |
- |
- |
- |
| 11 |
Hb_000349_130 |
0.2830230845 |
- |
- |
hypothetical protein JCGZ_06349 [Jatropha curcas] |
| 12 |
Hb_003850_030 |
0.283886565 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 13 |
Hb_001240_030 |
0.284401028 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 14 |
Hb_002326_160 |
0.2844520953 |
- |
- |
hypothetical protein JCGZ_04332 [Jatropha curcas] |
| 15 |
Hb_150093_010 |
0.2844763816 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 16 |
Hb_188247_020 |
0.284508038 |
- |
- |
hypothetical protein, partial [Cecembia lonarensis] |
| 17 |
Hb_000518_010 |
0.2845382714 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17300g [Populus trichocarpa] |
| 18 |
Hb_001900_040 |
0.2845555552 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 19 |
Hb_163720_010 |
0.2847824895 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 20 |
Hb_073174_010 |
0.2848902203 |
- |
- |
PREDICTED: MLP-like protein 328 [Jatropha curcas] |