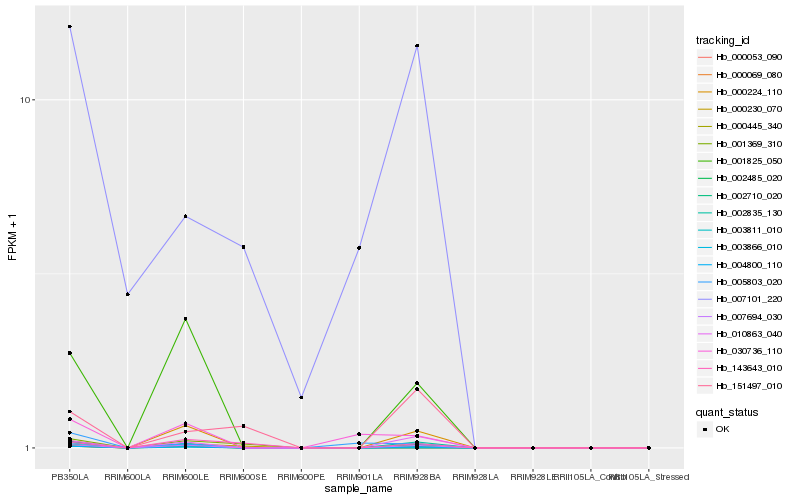
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002710_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_004800_110 |
0.0063099871 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK4 [Jatropha curcas] |
| 3 |
Hb_007694_030 |
0.0203958793 |
- |
- |
hypothetical protein SORBIDRAFT_04g037485 [Sorghum bicolor] |
| 4 |
Hb_000069_080 |
0.1306218736 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 5 |
Hb_003811_010 |
0.1330828246 |
- |
- |
PREDICTED: uncharacterized protein LOC102611294 [Citrus sinensis] |
| 6 |
Hb_002835_130 |
0.142851321 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 7 |
Hb_002485_020 |
0.1437159852 |
- |
- |
metal transport family protein [Populus trichocarpa] |
| 8 |
Hb_010863_040 |
0.1534701814 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 9 |
Hb_001825_050 |
0.1653220161 |
- |
- |
- |
| 10 |
Hb_000224_110 |
0.2474970017 |
- |
- |
- |
| 11 |
Hb_001369_310 |
0.3185356876 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_030736_110 |
0.3193831247 |
- |
- |
- |
| 13 |
Hb_143643_010 |
0.359384114 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC103967596 [Pyrus x bretschneideri] |
| 14 |
Hb_151497_010 |
0.3648112491 |
- |
- |
- |
| 15 |
Hb_005803_020 |
0.3655700694 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 16 |
Hb_007101_220 |
0.3750142416 |
- |
- |
Gibberellin-regulated protein 3 precursor, putative [Ricinus communis] |
| 17 |
Hb_000230_070 |
0.393036198 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 18 |
Hb_003866_010 |
0.4083662239 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 19 |
Hb_000445_340 |
0.4084391479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000053_090 |
0.408520414 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Populus euphratica] |