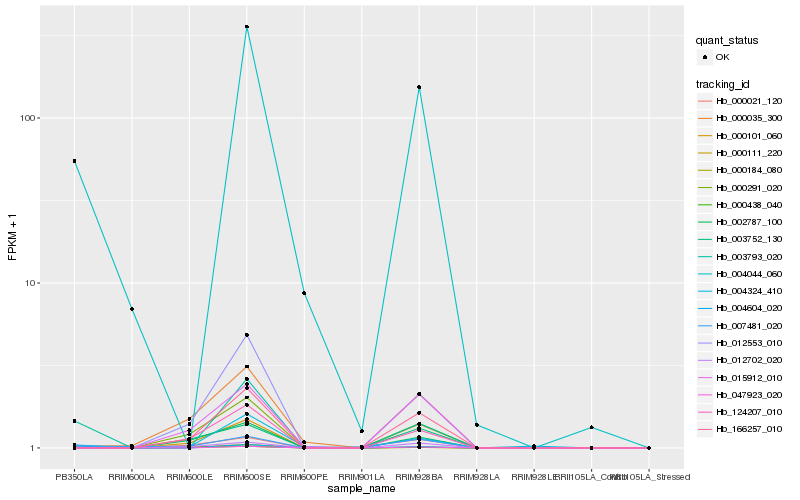
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_000184_080 |
0.1931114001 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 3 |
Hb_000021_120 |
0.2096448456 |
- |
- |
PREDICTED: uncharacterized protein LOC104115996 isoform X1 [Nicotiana tomentosiformis] |
| 4 |
Hb_007481_020 |
0.2280325156 |
- |
- |
- |
| 5 |
Hb_047923_020 |
0.2316903142 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_000111_220 |
0.2418954046 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_004044_060 |
0.2530671034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003793_020 |
0.2550763993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000035_300 |
0.2551631481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000291_020 |
0.2624054183 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 11 |
Hb_166257_010 |
0.264184365 |
- |
- |
hypothetical protein JCGZ_21808 [Jatropha curcas] |
| 12 |
Hb_124207_010 |
0.2669075576 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_004604_020 |
0.2670501112 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 14 |
Hb_002787_100 |
0.2696678931 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 15 |
Hb_012702_020 |
0.2716270953 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 16 |
Hb_015912_010 |
0.2732557798 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000101_060 |
0.2741305289 |
- |
- |
- |
| 18 |
Hb_003752_130 |
0.2759139052 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 19 |
Hb_004324_410 |
0.2771917928 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 20 |
Hb_012553_010 |
0.2794918306 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |