| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
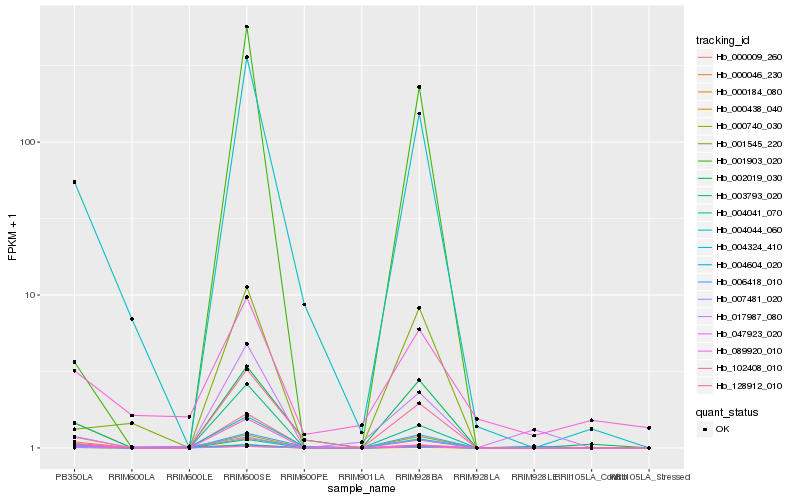
Hb_047923_020 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_004604_020 |
0.0993220534 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 3 |
Hb_003793_020 |
0.1415675511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004044_060 |
0.1803762237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000438_040 |
0.2316903142 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_004041_070 |
0.2362352618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000009_260 |
0.2378498584 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001545_220 |
0.2930946412 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 9 |
Hb_000740_030 |
0.2960659323 |
- |
- |
hypothetical protein VITISV_038271 [Vitis vinifera] |
| 10 |
Hb_128912_010 |
0.2975404544 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 11 |
Hb_004324_410 |
0.3065225752 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 12 |
Hb_007481_020 |
0.3093169203 |
- |
- |
- |
| 13 |
Hb_017987_080 |
0.3098043807 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 14 |
Hb_000046_230 |
0.3130838314 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 15 |
Hb_102408_010 |
0.3149091141 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 16 |
Hb_000184_080 |
0.3166969026 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 17 |
Hb_089920_010 |
0.3188147696 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_001903_020 |
0.3229247869 |
- |
- |
- |
| 19 |
Hb_002019_030 |
0.3231231739 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_006418_010 |
0.325457135 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |