| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004604_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 2 |
Hb_047923_020 |
0.0993220534 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
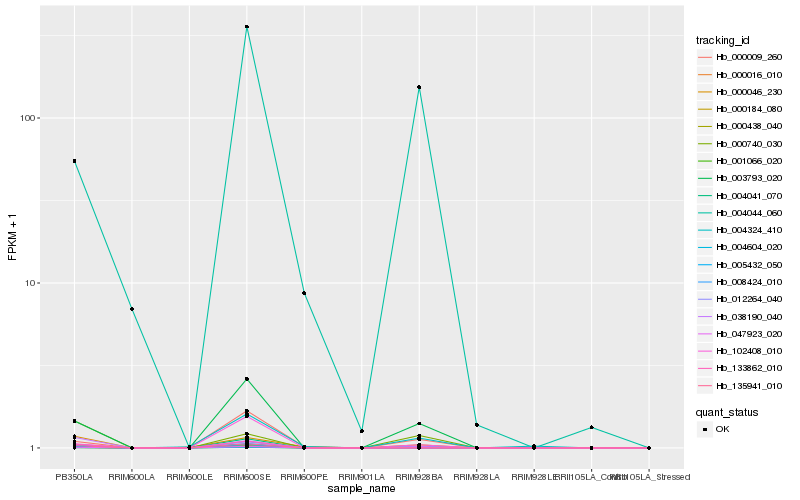
Hb_003793_020 |
0.1211476808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004041_070 |
0.2134507747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004044_060 |
0.2331682577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000009_260 |
0.2407208788 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000046_230 |
0.2449694819 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 8 |
Hb_000740_030 |
0.2463463188 |
- |
- |
hypothetical protein VITISV_038271 [Vitis vinifera] |
| 9 |
Hb_038190_040 |
0.2546973132 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 10 |
Hb_000438_040 |
0.2670501112 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_102408_010 |
0.2998999759 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 12 |
Hb_000184_080 |
0.3005131895 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 13 |
Hb_004324_410 |
0.3317214345 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 14 |
Hb_133862_010 |
0.3320656003 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 15 |
Hb_135941_010 |
0.3320915475 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 16 |
Hb_001066_020 |
0.3323494654 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_012264_040 |
0.3324189641 |
- |
- |
- |
| 18 |
Hb_005432_050 |
0.3339145644 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 19 |
Hb_008424_010 |
0.3387467709 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_000016_010 |
0.3389899734 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |