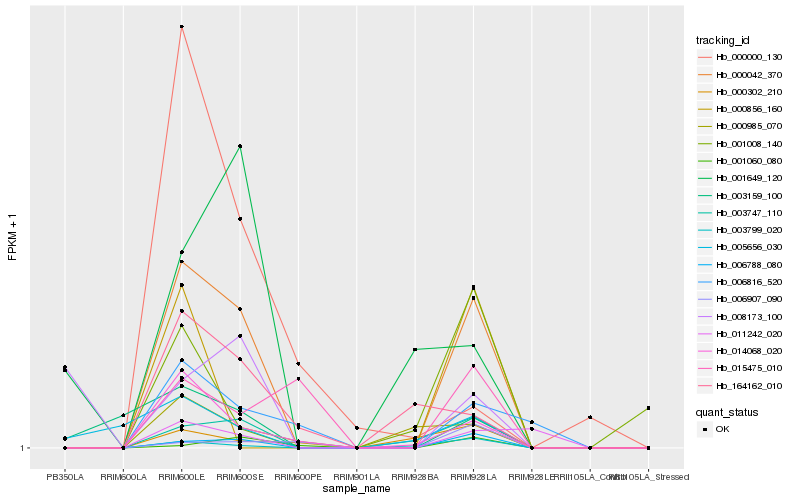
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_370 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006788_080 |
0.1918734838 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 3 |
Hb_003747_110 |
0.2391500264 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 4 |
Hb_014068_020 |
0.2679930697 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 5 |
Hb_003799_020 |
0.3037027814 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 6 |
Hb_006816_520 |
0.349144014 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 7 |
Hb_000985_070 |
0.3610469809 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 8 |
Hb_164162_010 |
0.368887386 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 9 |
Hb_003159_100 |
0.3691122737 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001060_080 |
0.3754942257 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 11 |
Hb_011242_020 |
0.3762035882 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 12 |
Hb_001008_140 |
0.390177252 |
- |
- |
- |
| 13 |
Hb_000302_210 |
0.3953848304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005656_030 |
0.3972994585 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001649_120 |
0.3980966944 |
- |
- |
- |
| 16 |
Hb_000856_160 |
0.3998202964 |
- |
- |
- |
| 17 |
Hb_015475_010 |
0.4100857269 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 18 |
Hb_000000_130 |
0.4106105897 |
- |
- |
Disease resistance-responsive family protein [Theobroma cacao] |
| 19 |
Hb_006907_090 |
0.4130015343 |
- |
- |
- |
| 20 |
Hb_008173_100 |
0.4156741256 |
- |
- |
Kiwellin, putative [Ricinus communis] |