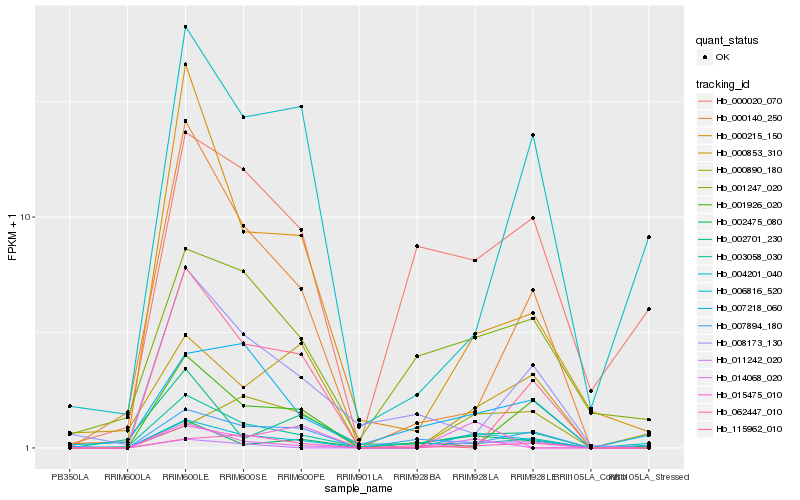
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_520 |
0.0 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 2 |
Hb_011242_020 |
0.2670455173 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_062447_010 |
0.2809237664 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 4 |
Hb_002475_080 |
0.285087633 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 5 |
Hb_014068_020 |
0.2924922353 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 6 |
Hb_001247_020 |
0.301589969 |
- |
- |
galactinol synthase family protein [Populus trichocarpa] |
| 7 |
Hb_007218_060 |
0.3096986878 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000140_250 |
0.3097559705 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 9 |
Hb_000215_150 |
0.3110116517 |
- |
- |
PREDICTED: uncharacterized protein LOC100249186 [Vitis vinifera] |
| 10 |
Hb_003058_030 |
0.3143897568 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000890_180 |
0.3172135255 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 12 |
Hb_000020_070 |
0.318545109 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 13 |
Hb_000853_310 |
0.3189627387 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 14 |
Hb_007894_180 |
0.3196373622 |
- |
- |
PREDICTED: cytochrome P450 716B1-like [Jatropha curcas] |
| 15 |
Hb_001926_020 |
0.330374933 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 16 |
Hb_015475_010 |
0.3335264772 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 17 |
Hb_004201_040 |
0.3349608688 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 18 |
Hb_002701_230 |
0.3353032094 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |
| 19 |
Hb_008173_130 |
0.3356866393 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 20 |
Hb_115962_010 |
0.3357693821 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |