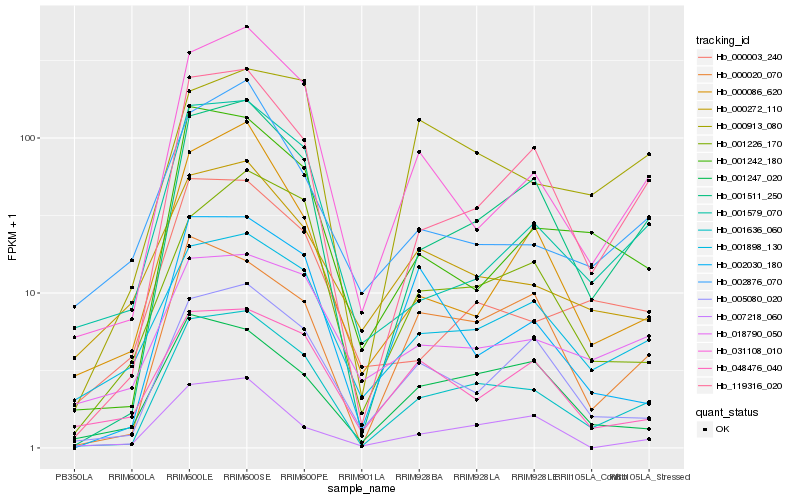
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001636_060 |
0.0 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 2 |
Hb_001511_250 |
0.0863680562 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 3 |
Hb_119316_020 |
0.1104537494 |
- |
- |
- |
| 4 |
Hb_031108_010 |
0.1437855125 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 5 |
Hb_000020_070 |
0.1478809366 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 6 |
Hb_005080_020 |
0.1519064081 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 7 |
Hb_001898_130 |
0.1521065365 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_001242_180 |
0.1651071315 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 9 |
Hb_001579_070 |
0.1687640898 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 10 |
Hb_001226_170 |
0.169256549 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_001247_020 |
0.1743142954 |
- |
- |
galactinol synthase family protein [Populus trichocarpa] |
| 12 |
Hb_007218_060 |
0.1746624854 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000003_240 |
0.1761075391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000913_080 |
0.178413198 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 15 |
Hb_018790_050 |
0.1795224298 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 16 |
Hb_000272_110 |
0.1834938921 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000086_620 |
0.1835296825 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_048476_040 |
0.184016677 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 19 |
Hb_002030_180 |
0.1851162218 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_002876_070 |
0.1873859001 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |