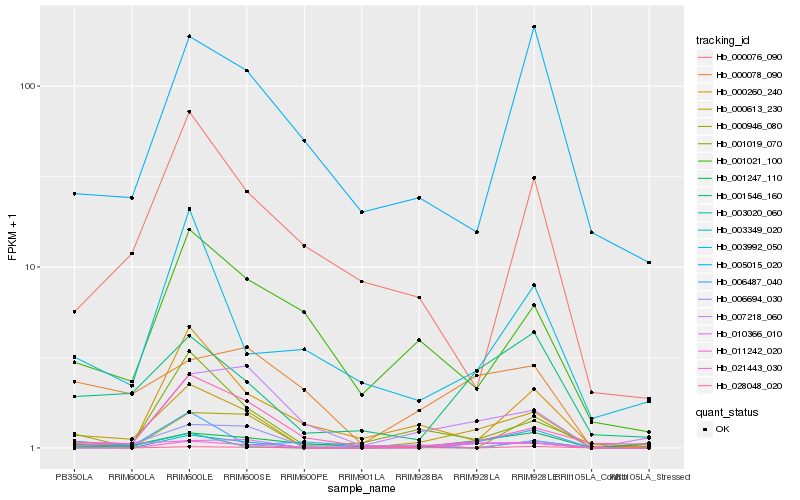
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_230 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010366_010 |
0.2550765886 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_003020_060 |
0.2638856345 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 4 |
Hb_001546_160 |
0.2688980378 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001019_070 |
0.2811097237 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 6 |
Hb_021443_030 |
0.2896032629 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 7 |
Hb_011242_020 |
0.2957633769 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_001247_110 |
0.3044673115 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 9 |
Hb_006694_030 |
0.3054976322 |
- |
- |
- |
| 10 |
Hb_006487_040 |
0.3147923642 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 11 |
Hb_000946_080 |
0.3170665449 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Prunus mume] |
| 12 |
Hb_000076_090 |
0.3186513302 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_003992_050 |
0.3192031414 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 14 |
Hb_000260_240 |
0.3193389878 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 15 |
Hb_028048_020 |
0.319367703 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 16 |
Hb_001021_100 |
0.3195827975 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 17 |
Hb_000078_090 |
0.3215297181 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 18 |
Hb_007218_060 |
0.3216897754 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003349_020 |
0.3230727022 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_005015_020 |
0.3240186129 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |