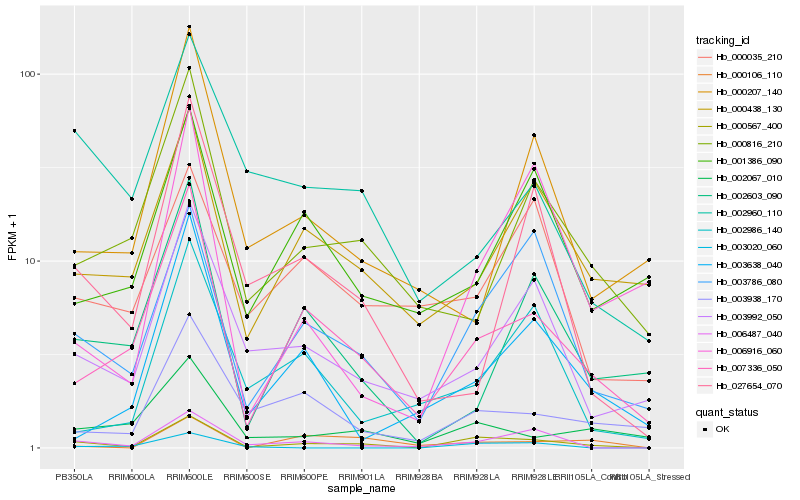
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_400 |
0.0 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 2 |
Hb_007336_050 |
0.2183606547 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 3 |
Hb_003786_080 |
0.2285403691 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 4 |
Hb_006487_040 |
0.2396167401 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 5 |
Hb_003992_050 |
0.2632191528 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 6 |
Hb_002067_010 |
0.270200112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000438_130 |
0.2732073358 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 8 |
Hb_003938_170 |
0.2733314433 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 9 |
Hb_000816_210 |
0.2796667657 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 10 |
Hb_002603_090 |
0.2904104908 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 11 |
Hb_027654_070 |
0.29091705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002960_110 |
0.2951936242 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001386_090 |
0.3039674958 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 14 |
Hb_000035_210 |
0.3050308855 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 15 |
Hb_003638_040 |
0.3053237106 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_002986_140 |
0.3057257667 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 17 |
Hb_000106_110 |
0.3073704618 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 18 |
Hb_006916_060 |
0.3088870043 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 19 |
Hb_003020_060 |
0.3095081494 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 20 |
Hb_000207_140 |
0.3110220697 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |