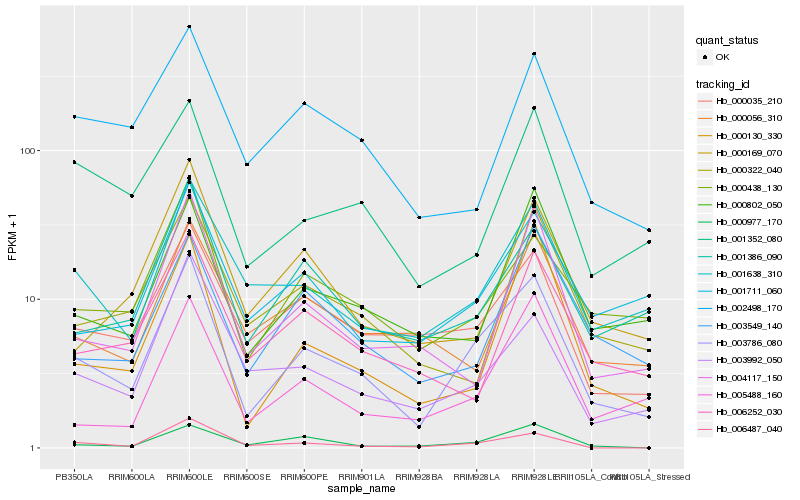
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003786_080 |
0.0 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 2 |
Hb_000035_210 |
0.1667307216 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 3 |
Hb_001638_310 |
0.1748840057 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_000438_130 |
0.1763124824 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 5 |
Hb_000977_170 |
0.1772589881 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_006487_040 |
0.1792023878 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 7 |
Hb_000802_050 |
0.1795348206 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 8 |
Hb_001386_090 |
0.1795481913 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 9 |
Hb_001711_060 |
0.181647189 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 10 |
Hb_005488_160 |
0.18505725 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_003549_140 |
0.1878159025 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_003992_050 |
0.1880808957 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 13 |
Hb_004117_150 |
0.1899324671 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 14 |
Hb_002498_170 |
0.190030215 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001352_080 |
0.1910036831 |
- |
- |
PREDICTED: uncharacterized protein At5g02240 [Jatropha curcas] |
| 16 |
Hb_000130_330 |
0.1910056339 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 17 |
Hb_000169_070 |
0.195794612 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 18 |
Hb_000322_040 |
0.1959882318 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006252_030 |
0.1973575577 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 20 |
Hb_000056_310 |
0.1987149541 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |