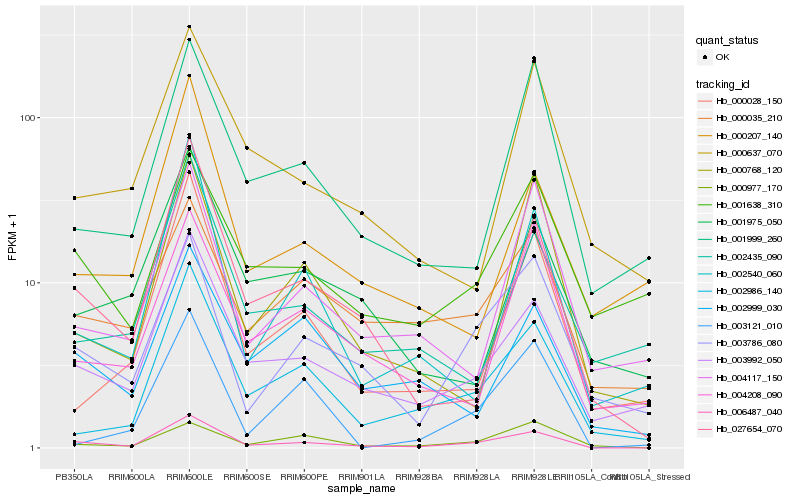
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006487_040 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 2 |
Hb_003992_050 |
0.1582013588 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 3 |
Hb_027654_070 |
0.1762851139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003786_080 |
0.1792023878 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_002986_140 |
0.1890423159 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 6 |
Hb_002999_030 |
0.2064432009 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 7 |
Hb_000035_210 |
0.2084979943 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 8 |
Hb_002540_060 |
0.2086790744 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001999_260 |
0.2142907662 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000028_150 |
0.2181878285 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 11 |
Hb_000768_120 |
0.2190886675 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 12 |
Hb_004208_090 |
0.219478117 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 13 |
Hb_000977_170 |
0.21982744 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_004117_150 |
0.2228259421 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 15 |
Hb_001638_310 |
0.2232308771 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 16 |
Hb_001975_050 |
0.2260387803 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 17 |
Hb_000637_070 |
0.2266040574 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 18 |
Hb_002435_090 |
0.2305028663 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000207_140 |
0.2326148296 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 20 |
Hb_003121_010 |
0.2350080369 |
- |
- |
hypothetical protein POPTR_0018s09720g [Populus trichocarpa] |