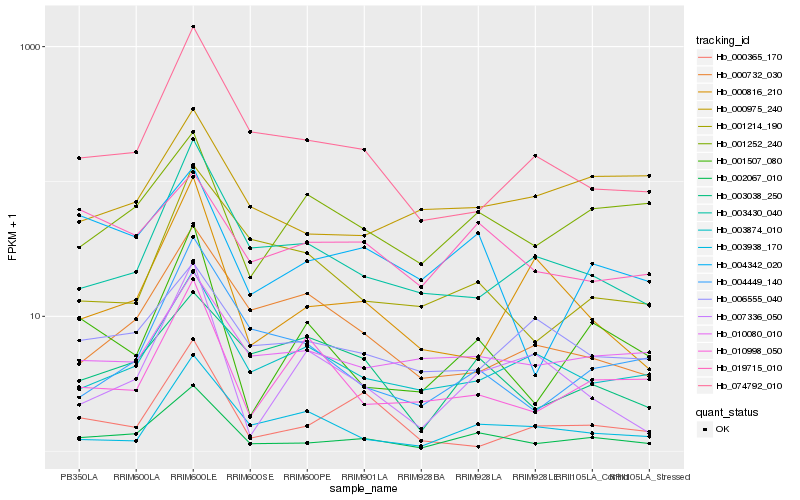
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002067_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001507_080 |
0.1815744134 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 3 |
Hb_074792_010 |
0.1936607041 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 4 |
Hb_003430_040 |
0.1956510304 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 5 |
Hb_003874_010 |
0.2006276523 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_004342_020 |
0.20706779 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_010080_010 |
0.2097647115 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 8 |
Hb_004449_140 |
0.21170647 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 9 |
Hb_001252_240 |
0.211853783 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 10 |
Hb_003938_170 |
0.2222338427 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 11 |
Hb_001214_190 |
0.2232643713 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 12 |
Hb_000816_210 |
0.2236863765 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 13 |
Hb_007336_050 |
0.2255241068 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 14 |
Hb_010998_050 |
0.2271671688 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003038_250 |
0.2308321995 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 16 |
Hb_000365_170 |
0.2311128824 |
- |
- |
PREDICTED: 50S ribosomal protein L17, chloroplastic isoform X3 [Jatropha curcas] |
| 17 |
Hb_019715_010 |
0.2330888193 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 18 |
Hb_000975_240 |
0.2337665282 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 19 |
Hb_006555_040 |
0.23404304 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 20 |
Hb_000732_030 |
0.2341120503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |