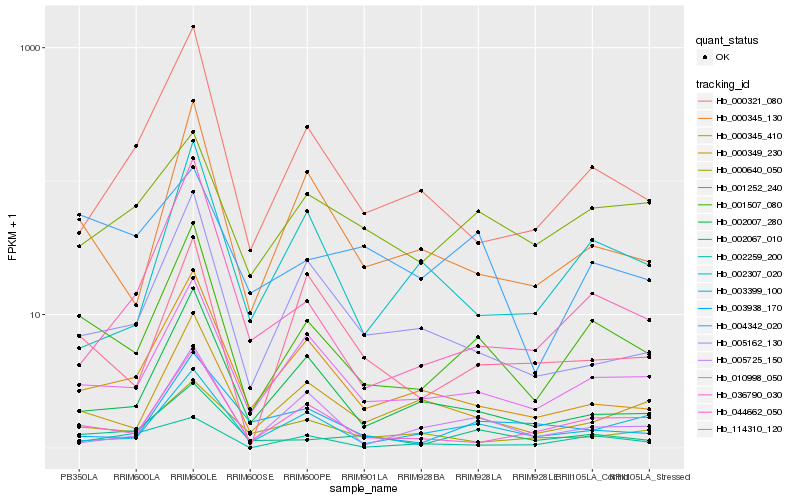
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001507_080 |
0.0 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 2 |
Hb_010998_050 |
0.1519401658 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 3 |
Hb_036790_030 |
0.1745078152 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 4 |
Hb_000345_130 |
0.1761212986 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002067_010 |
0.1815744134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000349_230 |
0.1951863492 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 7 |
Hb_000345_410 |
0.1952297847 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 8 |
Hb_002007_280 |
0.1969602781 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 9 |
Hb_005162_130 |
0.2133809855 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000321_080 |
0.2156146653 |
- |
- |
histone H4 [Zea mays] |
| 11 |
Hb_000640_050 |
0.2196499135 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 12 |
Hb_003399_100 |
0.2234388296 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 13 |
Hb_044662_050 |
0.2241778084 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 14 |
Hb_114310_120 |
0.2242319626 |
- |
- |
PREDICTED: 50S ribosomal protein L31, chloroplastic [Eucalyptus grandis] |
| 15 |
Hb_001252_240 |
0.2352960243 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 16 |
Hb_005725_150 |
0.2354323765 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002307_020 |
0.2376296126 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 18 |
Hb_003938_170 |
0.2384884214 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 19 |
Hb_004342_020 |
0.2385683393 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002259_200 |
0.2412102177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |