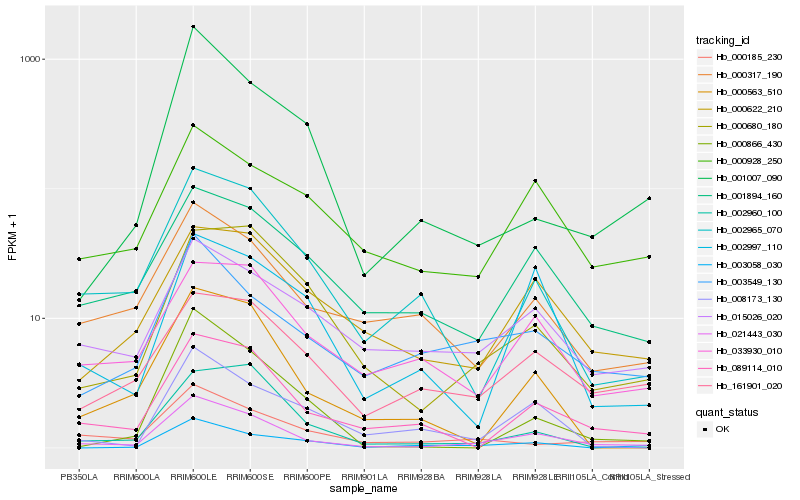
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021443_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 2 |
Hb_089114_010 |
0.1755201073 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 3 |
Hb_002965_070 |
0.1973034086 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 4 |
Hb_000680_180 |
0.1980302488 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 5 |
Hb_003549_130 |
0.1980395361 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 6 |
Hb_001894_160 |
0.2010444226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000563_510 |
0.201172118 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 8 |
Hb_000928_250 |
0.2071492408 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000317_190 |
0.2079292816 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 10 |
Hb_000622_210 |
0.2082449922 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_161901_020 |
0.2088412923 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 12 |
Hb_033930_010 |
0.209242573 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 13 |
Hb_008173_130 |
0.2094120233 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001007_090 |
0.2099246394 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 15 |
Hb_000866_430 |
0.2112122764 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 16 |
Hb_003058_030 |
0.2124038785 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000185_230 |
0.2125125435 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_015026_020 |
0.2128984364 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 19 |
Hb_002997_110 |
0.2134123102 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002960_100 |
0.2139855473 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |