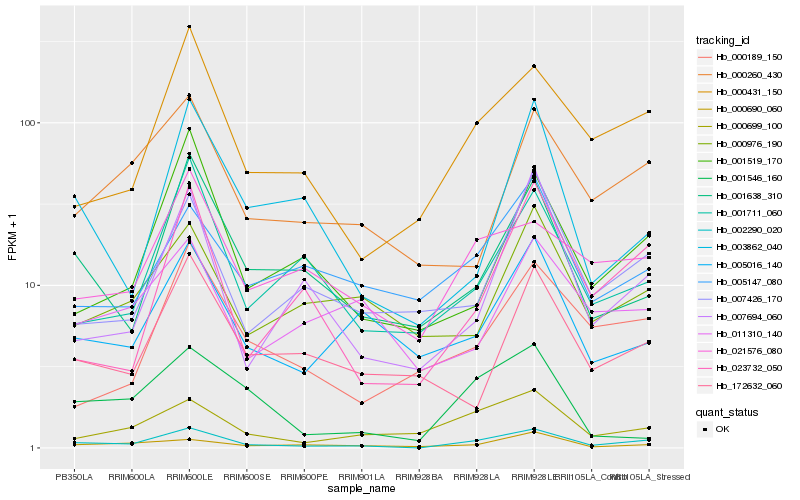
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002290_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000431_150 |
0.1847770336 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 3 |
Hb_000699_100 |
0.2089540467 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001638_310 |
0.2098096462 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 5 |
Hb_005016_140 |
0.2113126224 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_021576_080 |
0.2139238967 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 7 |
Hb_000260_430 |
0.2151700382 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 8 |
Hb_000690_060 |
0.2164183684 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 9 |
Hb_007426_170 |
0.2170114457 |
transcription factor |
TF Family: TCP |
- |
| 10 |
Hb_023732_050 |
0.2186577582 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 11 |
Hb_001711_060 |
0.2195746626 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 12 |
Hb_001519_170 |
0.2207914054 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007694_060 |
0.222249362 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000189_150 |
0.2264632294 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 15 |
Hb_005147_080 |
0.2274121662 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000976_190 |
0.2282983314 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 17 |
Hb_003862_040 |
0.232777227 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_172632_060 |
0.2335203235 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001546_160 |
0.2340714599 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_011310_140 |
0.2344155817 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |