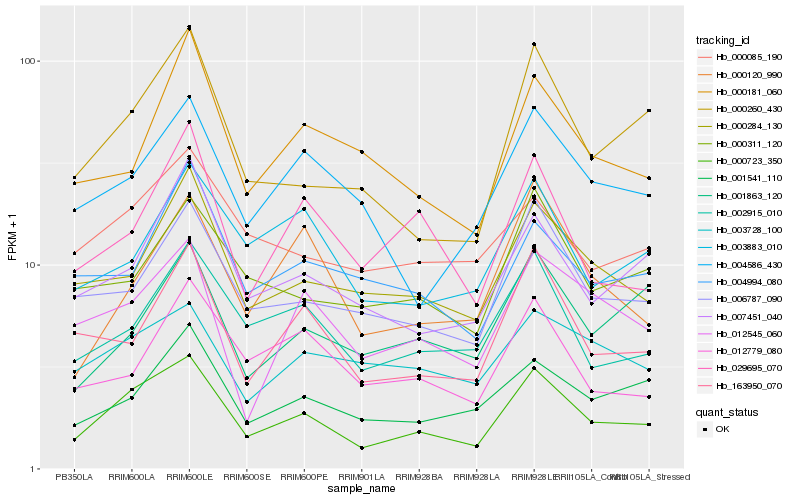
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_350 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000260_430 |
0.1320951647 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 3 |
Hb_006787_090 |
0.1345827546 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 4 |
Hb_002915_010 |
0.1383660479 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 5 |
Hb_000284_130 |
0.1383674092 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 6 |
Hb_163950_070 |
0.1395054492 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 7 |
Hb_007451_040 |
0.1417040314 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 8 |
Hb_001541_110 |
0.1447825324 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_012545_060 |
0.1459412813 |
- |
- |
PREDICTED: uncharacterized protein LOC103986927 [Musa acuminata subsp. malaccensis] |
| 10 |
Hb_000311_120 |
0.1517967131 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 11 |
Hb_029695_070 |
0.1524479857 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004586_430 |
0.1534742064 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003883_010 |
0.1541999901 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 14 |
Hb_000120_990 |
0.1560030403 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000085_190 |
0.1563309367 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 16 |
Hb_001863_120 |
0.1574274832 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000181_060 |
0.1592440612 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 18 |
Hb_012779_080 |
0.1594117383 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_003728_100 |
0.1604583784 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004994_080 |
0.1606040922 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |