| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
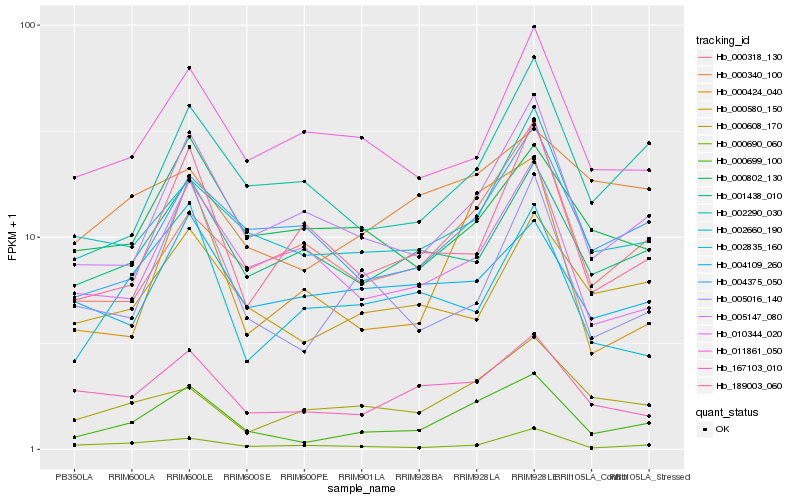
Hb_000699_100 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_005147_080 |
0.1609505053 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000424_040 |
0.1615644897 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 4 |
Hb_167103_010 |
0.1715782704 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 5 |
Hb_005016_140 |
0.1718434409 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_002660_190 |
0.1753766591 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 7 |
Hb_002290_030 |
0.1785512926 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000580_150 |
0.185456875 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_004375_050 |
0.1861658058 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000318_130 |
0.1878078624 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000608_170 |
0.1891200169 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 12 |
Hb_001438_010 |
0.1904600062 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000690_060 |
0.1925300927 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 14 |
Hb_010344_020 |
0.192677901 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000802_130 |
0.1948941527 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 16 |
Hb_189003_060 |
0.1971262608 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000340_100 |
0.1983550746 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002835_160 |
0.1992496538 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 19 |
Hb_011861_050 |
0.2002657828 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_004109_260 |
0.2022971541 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |