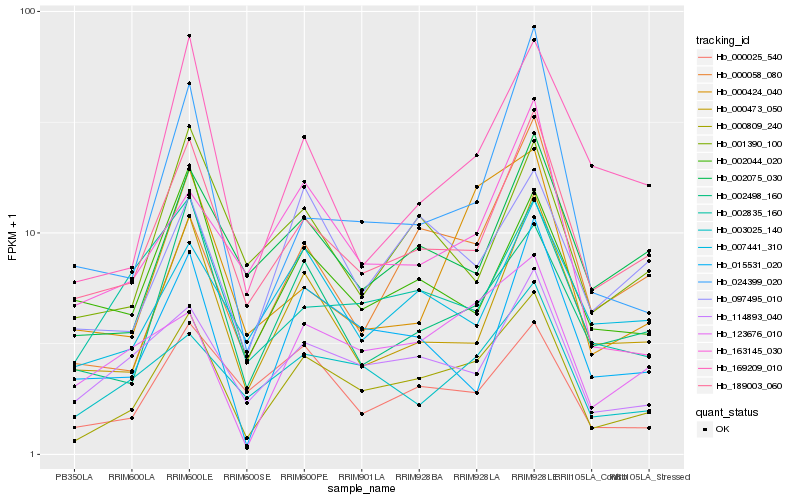
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_240 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 2 |
Hb_189003_060 |
0.1406574759 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_114893_040 |
0.151990715 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 4 |
Hb_000473_050 |
0.1590339682 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 5 |
Hb_002498_160 |
0.1629352339 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 6 |
Hb_123676_010 |
0.1655300949 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_003025_140 |
0.1660821379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000025_540 |
0.1739375122 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 9 |
Hb_169209_010 |
0.1751914897 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_015531_020 |
0.1759641716 |
- |
- |
PREDICTED: uncharacterized protein LOC100267182 isoform X2 [Vitis vinifera] |
| 11 |
Hb_002075_030 |
0.1762789976 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000058_080 |
0.1774133659 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_002835_160 |
0.1775558409 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 14 |
Hb_163145_030 |
0.1792122412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g46580, chloroplastic [Jatropha curcas] |
| 15 |
Hb_097495_010 |
0.18087725 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 16 |
Hb_024399_020 |
0.1812705064 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 17 |
Hb_007441_310 |
0.1813184475 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000424_040 |
0.1816568127 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 19 |
Hb_002044_020 |
0.1826397501 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 20 |
Hb_001390_100 |
0.1831917863 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |