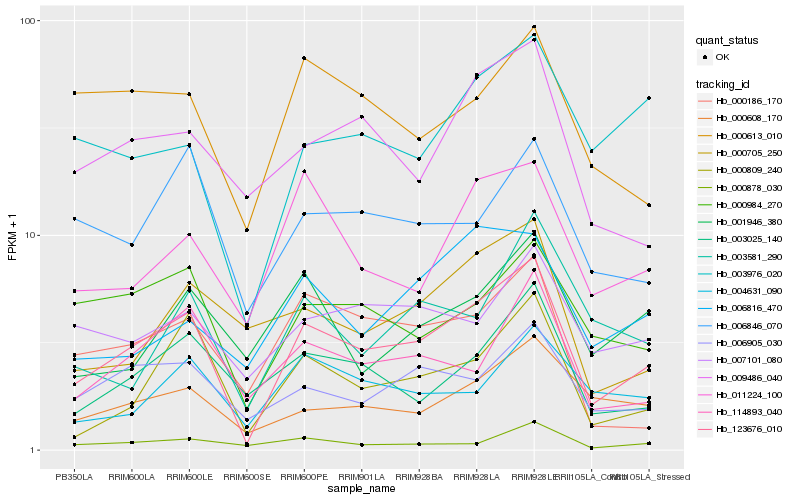
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123676_010 |
0.0 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_000809_240 |
0.1655300949 |
- |
- |
hypothetical protein POPTR_0007s11280g [Populus trichocarpa] |
| 3 |
Hb_006905_030 |
0.166763551 |
- |
- |
- |
| 4 |
Hb_000984_270 |
0.1697105047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000608_170 |
0.1711769696 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 6 |
Hb_114893_040 |
0.1715689466 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 7 |
Hb_003025_140 |
0.1745210902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011224_100 |
0.1788121178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009486_040 |
0.1802672909 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003976_020 |
0.1820297482 |
- |
- |
Regulator of ribonuclease activity A, putative [Ricinus communis] |
| 11 |
Hb_006846_070 |
0.1836719904 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_000186_170 |
0.1837817716 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 13 |
Hb_007101_080 |
0.1839377558 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 14 |
Hb_000878_030 |
0.1870942809 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 15 |
Hb_004631_090 |
0.1882055303 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 16 |
Hb_000613_010 |
0.1884210018 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 17 |
Hb_000705_250 |
0.1903084069 |
- |
- |
hypothetical protein CICLE_v10017607mg [Citrus clementina] |
| 18 |
Hb_003581_290 |
0.1913326047 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_006816_470 |
0.1926749179 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001946_380 |
0.1943688441 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |