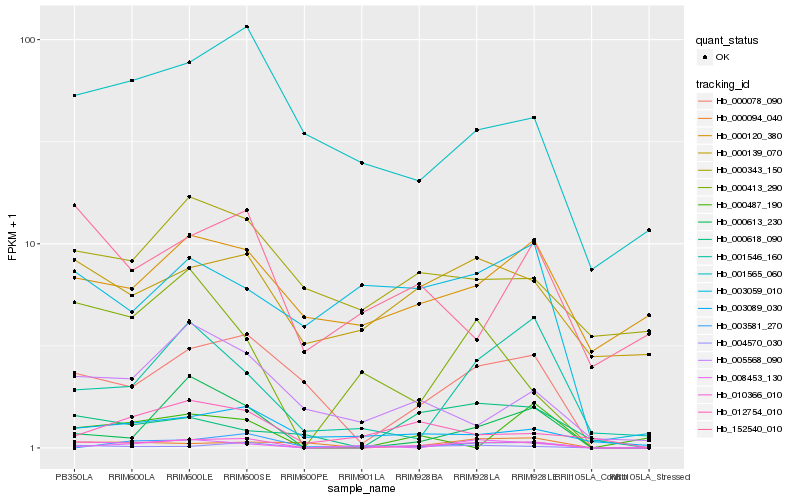
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010366_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_000078_090 |
0.2277290773 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 3 |
Hb_000613_230 |
0.2550765886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000413_290 |
0.2857694623 |
- |
- |
- |
| 5 |
Hb_001546_160 |
0.2974194157 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_003089_030 |
0.2978667036 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 7 |
Hb_005568_090 |
0.3028171142 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_001565_060 |
0.3044040593 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000094_040 |
0.3053771332 |
- |
- |
- |
| 10 |
Hb_152540_010 |
0.3154002064 |
- |
- |
- |
| 11 |
Hb_000139_070 |
0.3181942533 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 12 |
Hb_000618_090 |
0.3210966359 |
- |
- |
- |
| 13 |
Hb_004570_030 |
0.3263788234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012754_010 |
0.3276279186 |
- |
- |
- |
| 15 |
Hb_000343_150 |
0.3357764706 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 16 |
Hb_000487_190 |
0.3363689103 |
- |
- |
- |
| 17 |
Hb_003059_010 |
0.3378370574 |
- |
- |
PREDICTED: uncharacterized protein LOC104236538 [Nicotiana sylvestris] |
| 18 |
Hb_008453_130 |
0.3382452382 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 19 |
Hb_000120_380 |
0.3390349836 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 20 |
Hb_003581_270 |
0.3424927008 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic-like isoform X1 [Citrus sinensis] |