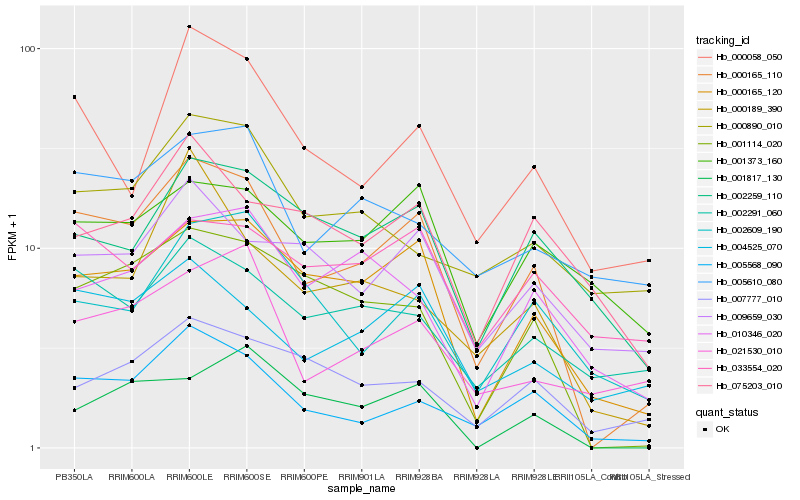
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_110 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_000165_120 |
0.1388945917 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 3 |
Hb_005568_090 |
0.1442638183 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_001114_020 |
0.1470608507 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 5 |
Hb_004525_070 |
0.1693293062 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_000058_050 |
0.1711041814 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 7 |
Hb_002291_060 |
0.1804507043 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_009659_030 |
0.1872135265 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 9 |
Hb_002259_110 |
0.1891281878 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 10 |
Hb_007777_010 |
0.1895656661 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 11 |
Hb_010346_020 |
0.190695212 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 12 |
Hb_021530_010 |
0.1926364198 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 13 |
Hb_002609_190 |
0.1950506489 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 14 |
Hb_005610_080 |
0.1953718018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000189_390 |
0.1961066474 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001373_160 |
0.1997652658 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 17 |
Hb_001817_130 |
0.2003682935 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 18 |
Hb_033554_020 |
0.2011150538 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 19 |
Hb_000890_010 |
0.2018880438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_075203_010 |
0.2021498735 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |