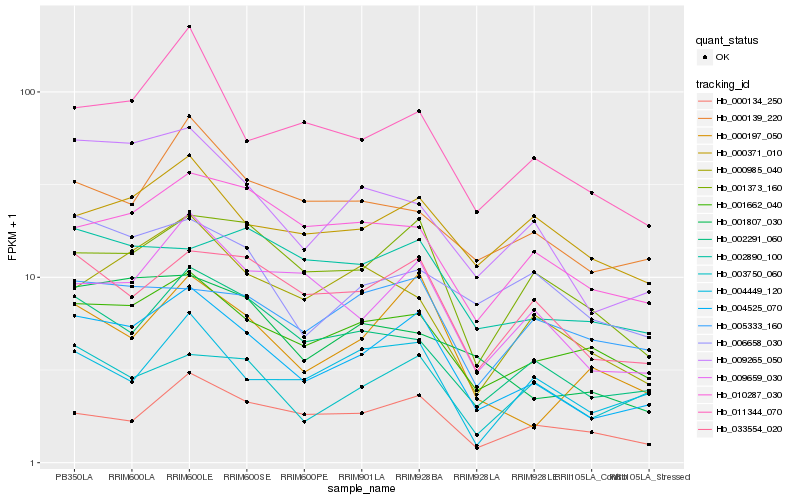
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_070 |
0.0 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 2 |
Hb_001662_040 |
0.1223593139 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 3 |
Hb_009265_050 |
0.1242056639 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 4 |
Hb_033554_020 |
0.1276038887 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 5 |
Hb_002291_060 |
0.1283556382 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_009659_030 |
0.1334476634 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 7 |
Hb_001373_160 |
0.1349181944 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 8 |
Hb_000197_050 |
0.1386967416 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 9 |
Hb_000134_250 |
0.1404558039 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_011344_070 |
0.141840498 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 11 |
Hb_003750_060 |
0.1496244173 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006658_030 |
0.1498101259 |
- |
- |
hypothetical protein JCGZ_04047 [Jatropha curcas] |
| 13 |
Hb_000139_220 |
0.1511179774 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001807_030 |
0.1520227408 |
- |
- |
PREDICTED: CDT1-like protein b [Jatropha curcas] |
| 15 |
Hb_010287_030 |
0.1527207883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000371_010 |
0.152885399 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 17 |
Hb_004449_120 |
0.1561472315 |
- |
- |
PREDICTED: uncharacterized protein LOC105631291 [Jatropha curcas] |
| 18 |
Hb_000985_040 |
0.157295591 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 19 |
Hb_005333_160 |
0.1574891304 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 20 |
Hb_002890_100 |
0.1601041817 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |