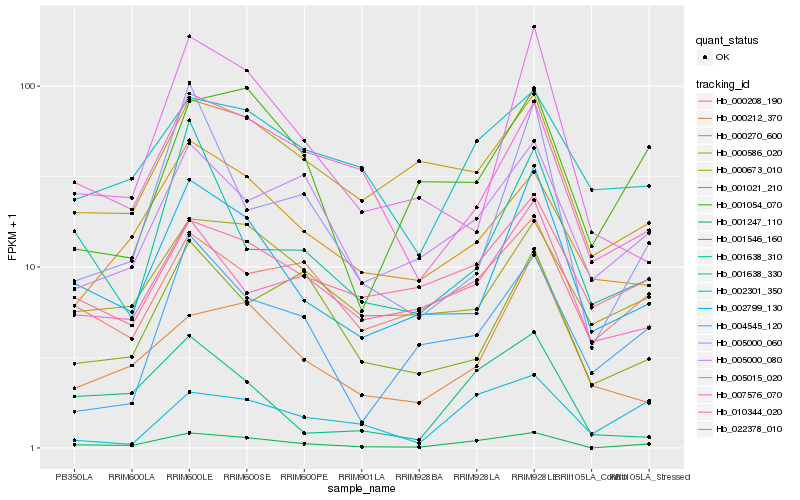
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_110 |
0.0 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 2 |
Hb_002799_130 |
0.1936308838 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 3 |
Hb_001546_160 |
0.2103004557 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_005000_080 |
0.2179626397 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 5 |
Hb_007576_070 |
0.2189406706 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000270_600 |
0.2198337675 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001054_070 |
0.2204846873 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 8 |
Hb_000208_190 |
0.2245237939 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 9 |
Hb_001638_330 |
0.2251039476 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000586_020 |
0.2264507938 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 11 |
Hb_004545_120 |
0.2267002599 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 12 |
Hb_005000_060 |
0.2283134951 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 13 |
Hb_000673_010 |
0.228437143 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000212_370 |
0.2293113633 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 15 |
Hb_010344_020 |
0.2302580963 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 16 |
Hb_005015_020 |
0.2336187705 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_002301_350 |
0.2340776643 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 18 |
Hb_001021_210 |
0.2340817317 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_022378_010 |
0.2346065837 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001638_310 |
0.235244417 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |