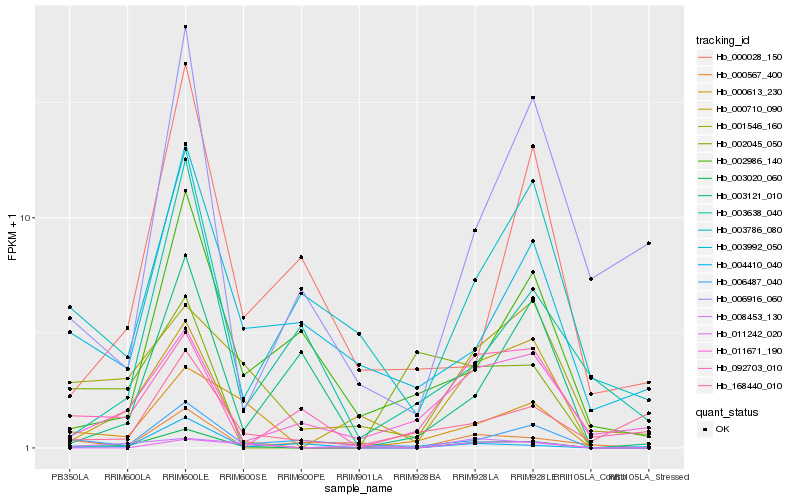
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_060 |
0.0 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 2 |
Hb_000613_230 |
0.2638856345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000710_090 |
0.2644428915 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 4 |
Hb_006487_040 |
0.2789981681 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 5 |
Hb_000567_400 |
0.3095081494 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 6 |
Hb_001546_160 |
0.3127767406 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_003992_050 |
0.3218729437 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 8 |
Hb_004410_040 |
0.3231592506 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_011242_020 |
0.3235172227 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_008453_130 |
0.3241207696 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 11 |
Hb_011671_190 |
0.3306612127 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 12 |
Hb_003786_080 |
0.3351764035 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 13 |
Hb_006916_060 |
0.3372135743 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 14 |
Hb_002986_140 |
0.3424943825 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 15 |
Hb_092703_010 |
0.3429735745 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 16 |
Hb_168440_010 |
0.3508842467 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 17 |
Hb_003121_010 |
0.3509319548 |
- |
- |
hypothetical protein POPTR_0018s09720g [Populus trichocarpa] |
| 18 |
Hb_002045_050 |
0.3521164822 |
- |
- |
- |
| 19 |
Hb_000028_150 |
0.3523431659 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 20 |
Hb_003638_040 |
0.3551302627 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |