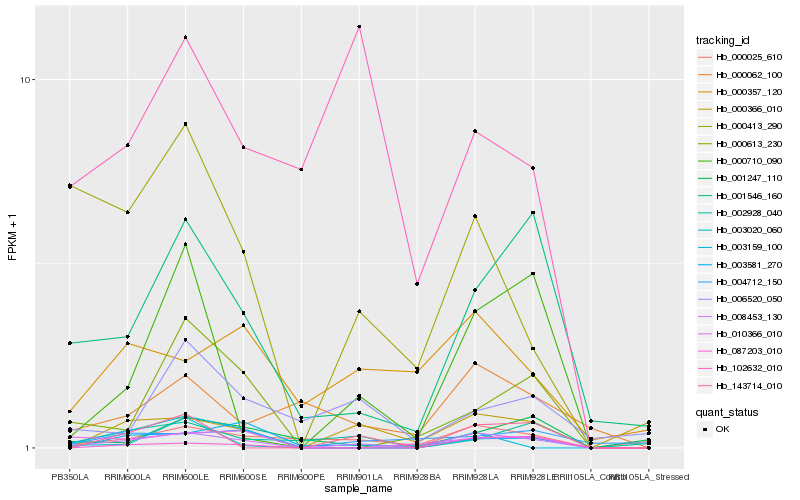
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008453_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 2 |
Hb_001546_160 |
0.2687852944 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000710_090 |
0.2944370888 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 4 |
Hb_000025_610 |
0.2983960457 |
- |
- |
annexin, putative [Ricinus communis] |
| 5 |
Hb_000413_290 |
0.2998883605 |
- |
- |
- |
| 6 |
Hb_143714_010 |
0.3092082718 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Populus euphratica] |
| 7 |
Hb_087203_010 |
0.3171211684 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Pyrus x bretschneideri] |
| 8 |
Hb_003020_060 |
0.3241207696 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 9 |
Hb_002928_040 |
0.3260687242 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 10 |
Hb_003581_270 |
0.3322669733 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_000062_100 |
0.3380868505 |
- |
- |
- |
| 12 |
Hb_010366_010 |
0.3382452382 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_000613_230 |
0.3389019752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000357_120 |
0.3394019526 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 15 |
Hb_000366_010 |
0.3447416822 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like, partial [Pyrus x bretschneideri] |
| 16 |
Hb_006520_050 |
0.352216183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_102632_010 |
0.3532782009 |
- |
- |
PREDICTED: CST complex subunit STN1 [Jatropha curcas] |
| 18 |
Hb_001247_110 |
0.3594266948 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_003159_100 |
0.3621137734 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004712_150 |
0.3624641216 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |