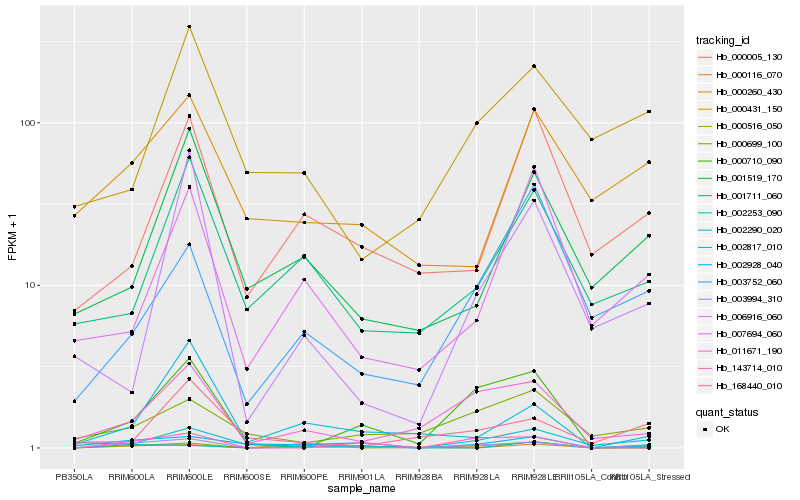
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_310 |
0.0 |
transcription factor |
TF Family: PHD |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011671_190 |
0.3048927876 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_143714_010 |
0.3145479685 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Populus euphratica] |
| 4 |
Hb_002290_020 |
0.3206968681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002928_040 |
0.3217451185 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 6 |
Hb_000710_090 |
0.3384729984 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 7 |
Hb_000516_050 |
0.338881272 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 8 |
Hb_001519_170 |
0.3453064716 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002817_010 |
0.3465904247 |
- |
- |
PREDICTED: uncharacterized protein LOC105630103 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000116_070 |
0.3483167212 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 11 |
Hb_000699_100 |
0.3488163973 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000260_430 |
0.3490223616 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 13 |
Hb_006916_060 |
0.3537721936 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 14 |
Hb_000431_150 |
0.3540059864 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 15 |
Hb_007694_060 |
0.3582514064 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002253_090 |
0.358251717 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_168440_010 |
0.3586461467 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 18 |
Hb_001711_060 |
0.358795464 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 19 |
Hb_000005_130 |
0.3592690427 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 20 |
Hb_003752_060 |
0.3596572462 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |