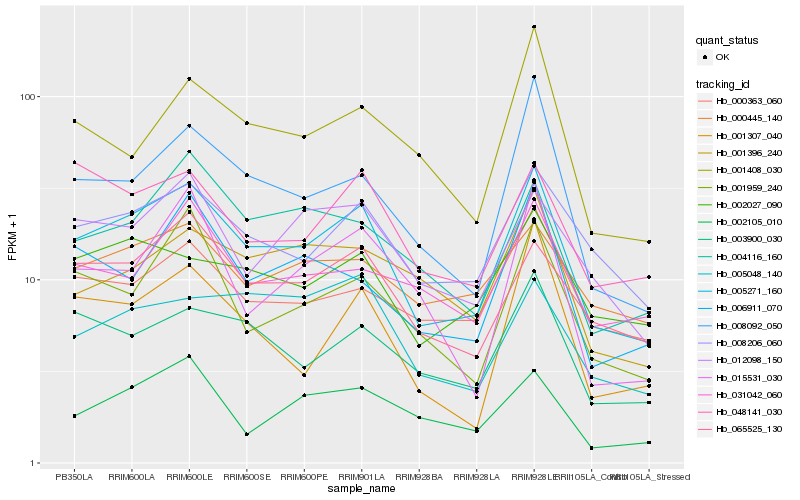
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 2 |
Hb_008206_060 |
0.1123870396 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_065525_130 |
0.1139656226 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_008092_050 |
0.1205472218 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 5 |
Hb_003900_030 |
0.1242380582 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012098_150 |
0.1308339193 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 7 |
Hb_001959_240 |
0.132438809 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_002027_090 |
0.1331192702 |
- |
- |
- |
| 9 |
Hb_004116_160 |
0.1334275636 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000363_060 |
0.1364720072 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006911_070 |
0.1367904461 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 12 |
Hb_048141_030 |
0.1388050293 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g59040-like [Prunus mume] |
| 13 |
Hb_001408_030 |
0.1397920844 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001396_240 |
0.1410671742 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002105_010 |
0.1412328638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001307_040 |
0.1419335981 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 4B [Jatropha curcas] |
| 17 |
Hb_000445_140 |
0.1431686118 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 18 |
Hb_031042_060 |
0.1438421454 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_005048_140 |
0.1441839433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_015531_030 |
0.1459307231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |