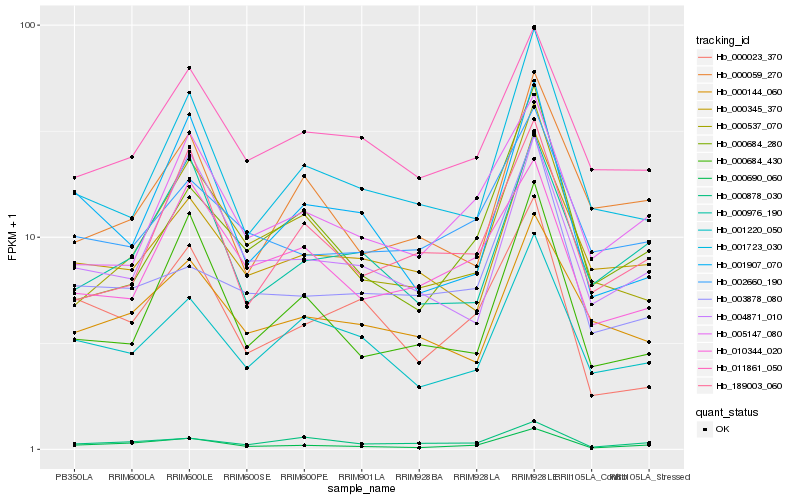
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000690_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 2 |
Hb_000976_190 |
0.1329439973 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 3 |
Hb_001220_050 |
0.1369306749 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 4 |
Hb_000878_030 |
0.1417448074 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001723_030 |
0.1424236179 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000023_370 |
0.1425564991 |
- |
- |
- |
| 7 |
Hb_000537_070 |
0.1445193918 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 8 |
Hb_001907_070 |
0.1450757999 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 9 |
Hb_002660_190 |
0.1455518187 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 10 |
Hb_005147_080 |
0.1455816191 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_004871_010 |
0.1472125408 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 12 |
Hb_011861_050 |
0.147652295 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_000684_430 |
0.1482311967 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000684_280 |
0.1515155608 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000345_370 |
0.1518918272 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 16 |
Hb_003878_080 |
0.1558588065 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 17 |
Hb_000144_060 |
0.1565576456 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_270 |
0.1569773753 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 19 |
Hb_010344_020 |
0.1577902729 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 20 |
Hb_189003_060 |
0.1583241903 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |