| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
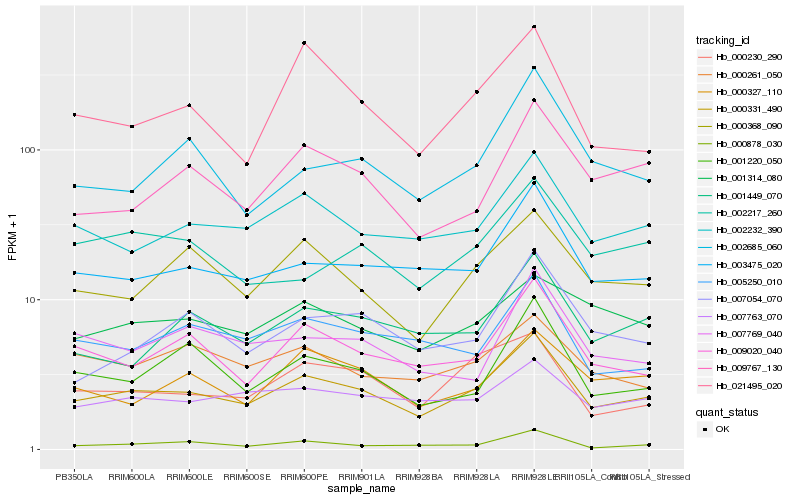
Hb_000331_490 |
0.0 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002232_390 |
0.0971448371 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 3 |
Hb_003475_020 |
0.0983164053 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 4 |
Hb_009020_040 |
0.1057703964 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000368_090 |
0.1121402036 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 6 |
Hb_000230_290 |
0.1179970413 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 7 |
Hb_000878_030 |
0.1194016466 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_009767_130 |
0.1194197305 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 9 |
Hb_001449_070 |
0.1201791869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007763_070 |
0.120338704 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005250_010 |
0.1220069747 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 12 |
Hb_001220_050 |
0.122845779 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 13 |
Hb_000327_110 |
0.1245959372 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 14 |
Hb_007054_070 |
0.1262817217 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 15 |
Hb_000261_050 |
0.1292759278 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 16 |
Hb_021495_020 |
0.12929161 |
- |
- |
chitinase, putative [Ricinus communis] |
| 17 |
Hb_002685_060 |
0.1305199409 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 18 |
Hb_007769_040 |
0.1310381773 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_001314_080 |
0.1318846968 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 20 |
Hb_002217_260 |
0.1320949073 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |