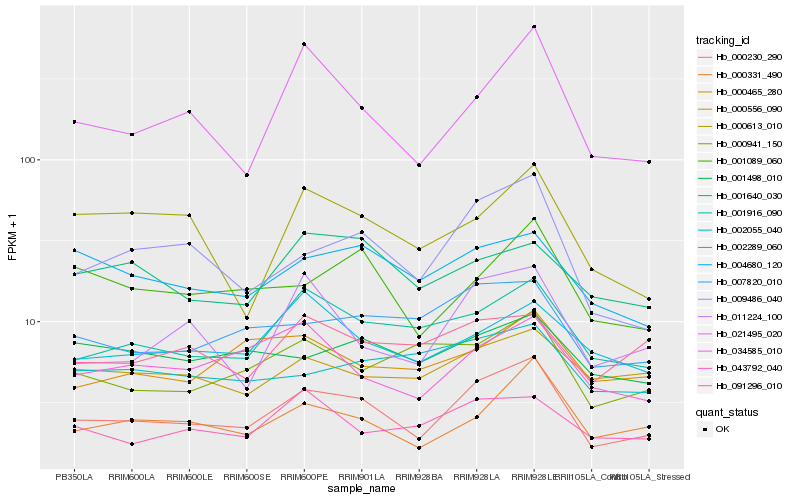
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_290 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_009486_040 |
0.10900552 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000331_490 |
0.1179970413 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001916_090 |
0.1226051778 |
- |
- |
beta-1,2-n-acetylglucosaminyltransferase II, putative [Ricinus communis] |
| 5 |
Hb_021495_020 |
0.1301326619 |
- |
- |
chitinase, putative [Ricinus communis] |
| 6 |
Hb_001089_060 |
0.1344457226 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1A isoform X1 [Jatropha curcas] |
| 7 |
Hb_007820_010 |
0.1348958582 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 8 |
Hb_011224_100 |
0.1401678482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_034585_010 |
0.1405471805 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 10 |
Hb_004680_120 |
0.1435111031 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 11 |
Hb_000941_150 |
0.1479861553 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002289_060 |
0.1496606707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000556_090 |
0.1500406514 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 14 |
Hb_043792_040 |
0.1506760716 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 15 |
Hb_000613_010 |
0.1540181654 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 16 |
Hb_000465_280 |
0.1541796851 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 17 |
Hb_001498_010 |
0.1569880241 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 18 |
Hb_001640_030 |
0.1570877653 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 19 |
Hb_091296_010 |
0.1571144302 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_002055_040 |
0.1571911817 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |