| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
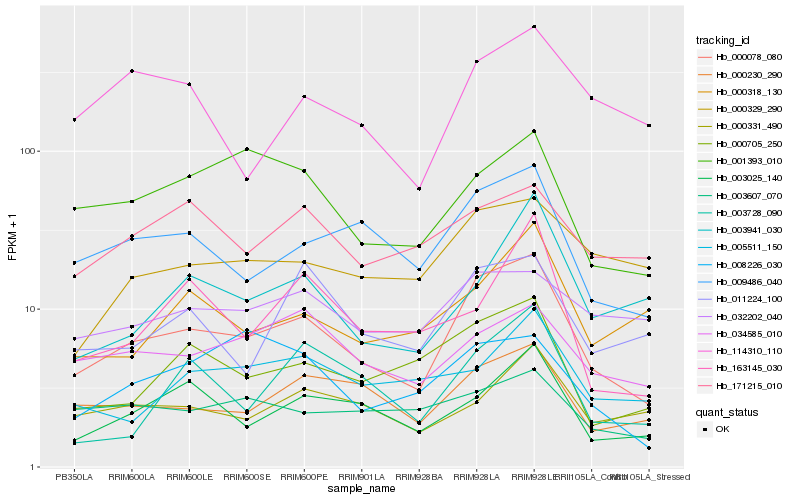
Hb_000078_080 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_17040 [Jatropha curcas] |
| 2 |
Hb_009486_040 |
0.1439944822 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000230_290 |
0.1573300493 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 4 |
Hb_000705_250 |
0.1609084006 |
- |
- |
hypothetical protein CICLE_v10017607mg [Citrus clementina] |
| 5 |
Hb_003025_140 |
0.1661214368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001393_010 |
0.1781777157 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 7 |
Hb_011224_100 |
0.1806124303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000329_290 |
0.1826237494 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 9 |
Hb_034585_010 |
0.1852881239 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 10 |
Hb_000318_130 |
0.1864127922 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000331_490 |
0.1866653371 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_114310_110 |
0.188196243 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_003941_030 |
0.1897551385 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 14 |
Hb_003728_090 |
0.1907210725 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 15 |
Hb_171215_010 |
0.1941810372 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 16 |
Hb_032202_040 |
0.1943441802 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 17 |
Hb_005511_150 |
0.1944340256 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |
| 18 |
Hb_003607_070 |
0.1949414231 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 19 |
Hb_163145_030 |
0.1951199088 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g46580, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008226_030 |
0.1964107026 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |