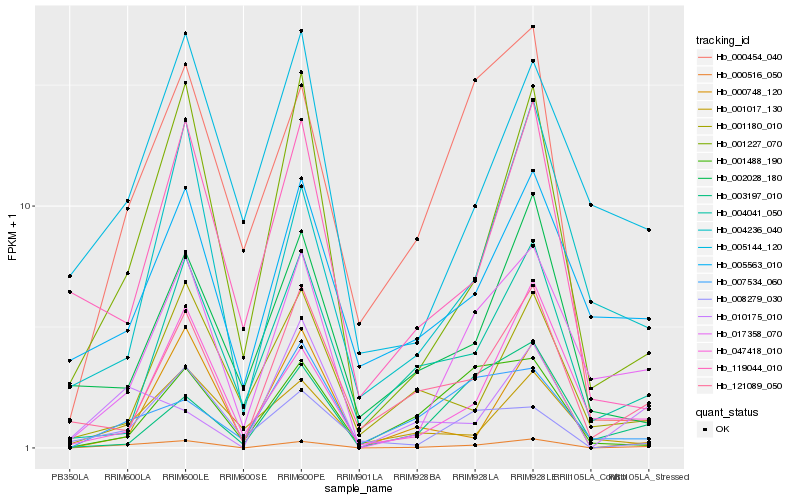
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000516_050 |
0.0 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 2 |
Hb_017358_070 |
0.2172896505 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 3 |
Hb_001227_070 |
0.2253290548 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 4 |
Hb_121089_050 |
0.2535877478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_047418_010 |
0.2539207366 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 6 |
Hb_000454_040 |
0.2554979846 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 7 |
Hb_004236_040 |
0.2631496153 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 8 |
Hb_005563_010 |
0.2679584429 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 9 |
Hb_004041_050 |
0.2707537019 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_001017_130 |
0.2710710991 |
- |
- |
hypothetical protein POPTR_0010s06070g [Populus trichocarpa] |
| 11 |
Hb_010175_010 |
0.2751437818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000748_120 |
0.2767551915 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 13 |
Hb_002028_180 |
0.2768766505 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_119044_010 |
0.2787645387 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_008279_030 |
0.2790571405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003197_010 |
0.2797863992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001488_190 |
0.2831679861 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 18 |
Hb_005144_120 |
0.2844790822 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 19 |
Hb_007534_060 |
0.2854068983 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_001180_010 |
0.2860088409 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |