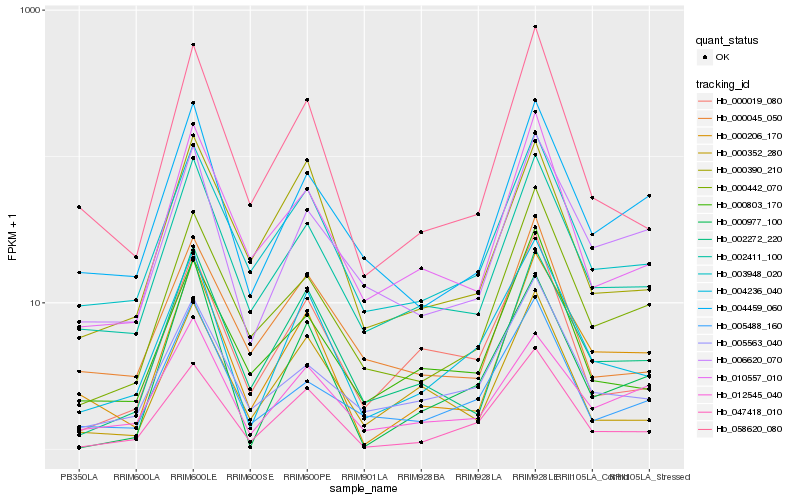
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004236_040 |
0.0 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 2 |
Hb_047418_010 |
0.0727267098 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 3 |
Hb_003948_020 |
0.1217195867 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 4 |
Hb_002411_100 |
0.1243573397 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 5 |
Hb_000019_080 |
0.126577385 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 6 |
Hb_058620_080 |
0.1315457429 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005563_040 |
0.1346316451 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_170 |
0.1454675148 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 9 |
Hb_006620_070 |
0.1485500852 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 10 |
Hb_010557_010 |
0.149313968 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002272_220 |
0.1495851792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005488_160 |
0.1502223526 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000352_280 |
0.1512792907 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 14 |
Hb_000206_170 |
0.151882688 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 15 |
Hb_000442_070 |
0.1534814819 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 16 |
Hb_004459_060 |
0.153568315 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000977_100 |
0.1541616964 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_012545_040 |
0.1573349929 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000390_210 |
0.15794771 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_000045_050 |
0.1581369973 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |