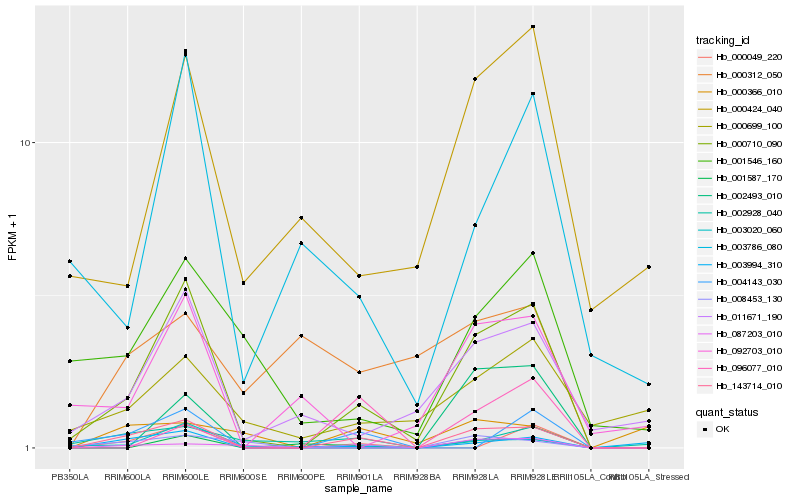
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143714_010 |
0.0 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Populus euphratica] |
| 2 |
Hb_000710_090 |
0.178187987 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 3 |
Hb_008453_130 |
0.3092082718 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 4 |
Hb_003994_310 |
0.3145479685 |
transcription factor |
TF Family: PHD |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011671_190 |
0.3514184215 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 6 |
Hb_001587_170 |
0.3582190479 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 7 |
Hb_000049_220 |
0.3608771192 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_003020_060 |
0.3629080219 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 9 |
Hb_002928_040 |
0.372248738 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 10 |
Hb_004143_030 |
0.3727002005 |
- |
- |
PREDICTED: chromosome-associated kinesin KIF4B [Jatropha curcas] |
| 11 |
Hb_087203_010 |
0.3798308326 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Pyrus x bretschneideri] |
| 12 |
Hb_096077_010 |
0.3876641513 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_003786_080 |
0.3897577707 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 14 |
Hb_001546_160 |
0.3933668119 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000699_100 |
0.3941248667 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01510, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000366_010 |
0.395103128 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like, partial [Pyrus x bretschneideri] |
| 17 |
Hb_000424_040 |
0.3972827 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 18 |
Hb_002493_010 |
0.3977542681 |
- |
- |
- |
| 19 |
Hb_092703_010 |
0.4021677241 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 20 |
Hb_000312_050 |
0.4056865707 |
- |
- |
transcription factor, putative [Ricinus communis] |