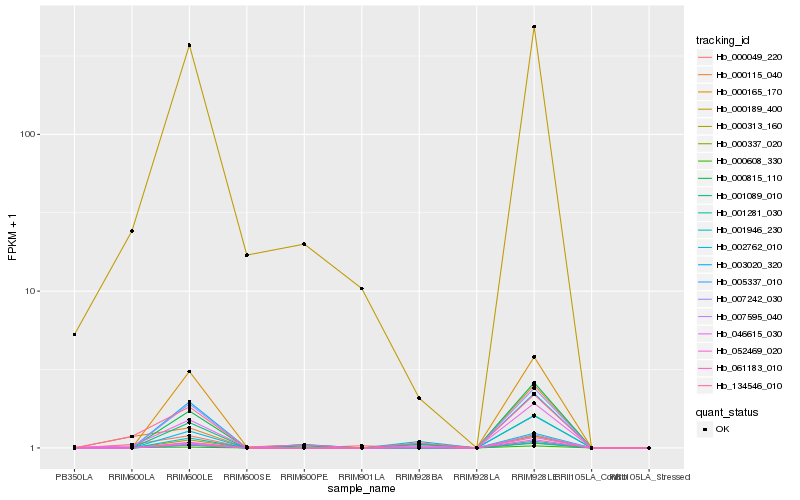
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134546_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 2 |
Hb_000608_330 |
0.1939739255 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 3 |
Hb_007242_030 |
0.2131207361 |
- |
- |
PREDICTED: methylesterase 3-like isoform X2 [Tarenaya hassleriana] |
| 4 |
Hb_046615_030 |
0.2197618548 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 5 |
Hb_000189_400 |
0.2266602255 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 6 |
Hb_000165_170 |
0.2293470384 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 7 |
Hb_001089_010 |
0.2337040673 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 8 |
Hb_000115_040 |
0.237932391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005337_010 |
0.2413141588 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 10 |
Hb_000815_110 |
0.243431924 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 11 |
Hb_000049_220 |
0.2437203934 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 12 |
Hb_002762_010 |
0.2437617143 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 13 |
Hb_061183_010 |
0.2438432233 |
- |
- |
PREDICTED: uncharacterized protein LOC105775353 [Gossypium raimondii] |
| 14 |
Hb_007595_040 |
0.2440660821 |
- |
- |
ribosomal protein L2 (chloroplast) [Berberis bealei] |
| 15 |
Hb_001946_230 |
0.2441438649 |
- |
- |
- |
| 16 |
Hb_000313_160 |
0.2442737837 |
- |
- |
BnaC02g20570D [Brassica napus] |
| 17 |
Hb_000337_020 |
0.2447538034 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001281_030 |
0.2449244592 |
- |
- |
hypothetical protein POPTR_0010s18450g [Populus trichocarpa] |
| 19 |
Hb_003020_320 |
0.2451457473 |
- |
- |
- |
| 20 |
Hb_052469_020 |
0.2452709577 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Vitis vinifera] |