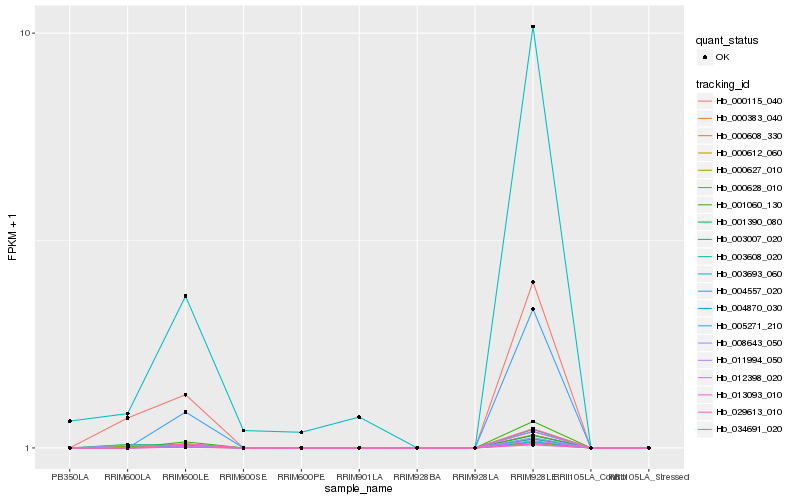
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000627_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 2 |
Hb_000115_040 |
0.020933825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000628_010 |
0.0997191818 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 4 |
Hb_001390_080 |
0.1031402121 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 5 |
Hb_008643_050 |
0.1036049597 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 6 |
Hb_000608_330 |
0.1311233255 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 7 |
Hb_003693_060 |
0.2199629576 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 8 |
Hb_012398_020 |
0.2258541367 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 9 |
Hb_013093_010 |
0.2258543835 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 10 |
Hb_004557_020 |
0.2258546507 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 11 |
Hb_029613_010 |
0.2258563176 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_000383_040 |
0.2258631478 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 13 |
Hb_003007_020 |
0.2258633878 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 14 |
Hb_000612_060 |
0.2258642417 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 15 |
Hb_034691_020 |
0.2258692027 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 16 |
Hb_003608_020 |
0.2258767116 |
- |
- |
hypothetical protein CISIN_1g041945mg [Citrus sinensis] |
| 17 |
Hb_004870_030 |
0.2259073005 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_011994_050 |
0.2259365896 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 19 |
Hb_001060_130 |
0.2260177965 |
- |
- |
- |
| 20 |
Hb_005271_210 |
0.2261604273 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |