| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
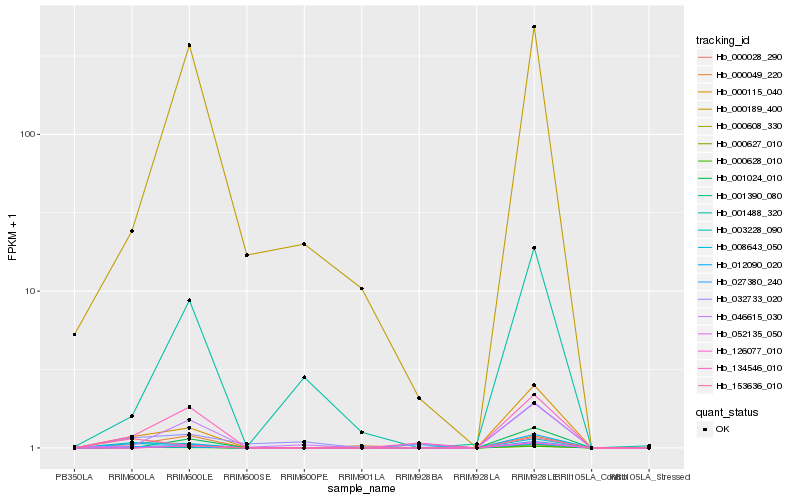
Hb_000608_330 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_001390_080 |
0.1006019489 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 3 |
Hb_000628_010 |
0.1056395486 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 4 |
Hb_008643_050 |
0.109913067 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 5 |
Hb_003228_090 |
0.1263652801 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 6 |
Hb_000115_040 |
0.1285853975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000627_010 |
0.1311233255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 8 |
Hb_134546_010 |
0.1939739255 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 9 |
Hb_153636_010 |
0.2561701995 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 10 |
Hb_000049_220 |
0.2653101508 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 11 |
Hb_012090_020 |
0.2728896503 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_046615_030 |
0.2791069539 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 13 |
Hb_032733_020 |
0.2831988611 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 14 |
Hb_027380_240 |
0.2843783754 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001488_320 |
0.2887200879 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_000189_400 |
0.2887557654 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 17 |
Hb_126077_010 |
0.2910593328 |
- |
- |
- |
| 18 |
Hb_001024_010 |
0.2910895103 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 19 |
Hb_000028_290 |
0.2910917076 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 20 |
Hb_052135_050 |
0.291200417 |
- |
- |
PREDICTED: probable pectinesterase 67 [Jatropha curcas] |