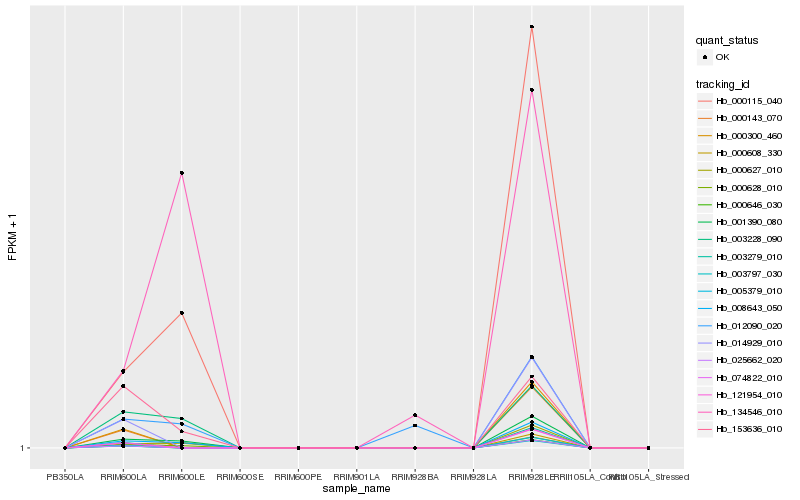
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001390_080 |
0.0 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 2 |
Hb_000628_010 |
0.0067411698 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 3 |
Hb_008643_050 |
0.0095253101 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 4 |
Hb_000608_330 |
0.1006019489 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 5 |
Hb_000627_010 |
0.1031402121 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 6 |
Hb_000115_040 |
0.1169326476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003228_090 |
0.1548135263 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 8 |
Hb_153636_010 |
0.2168767509 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 9 |
Hb_012090_020 |
0.2614070073 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_134546_010 |
0.2702164973 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 11 |
Hb_000300_460 |
0.2754514334 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 12 |
Hb_003279_010 |
0.2754625546 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 13 |
Hb_121954_010 |
0.2754650704 |
- |
- |
PREDICTED: uncharacterized protein LOC105774125 [Gossypium raimondii] |
| 14 |
Hb_074822_010 |
0.2754658347 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 15 |
Hb_005379_010 |
0.2754888139 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_000646_030 |
0.2754977667 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_025662_020 |
0.2754983565 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_003797_030 |
0.2755008269 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_000143_070 |
0.2755397689 |
- |
- |
- |
| 20 |
Hb_014929_010 |
0.2758548155 |
- |
- |
- |