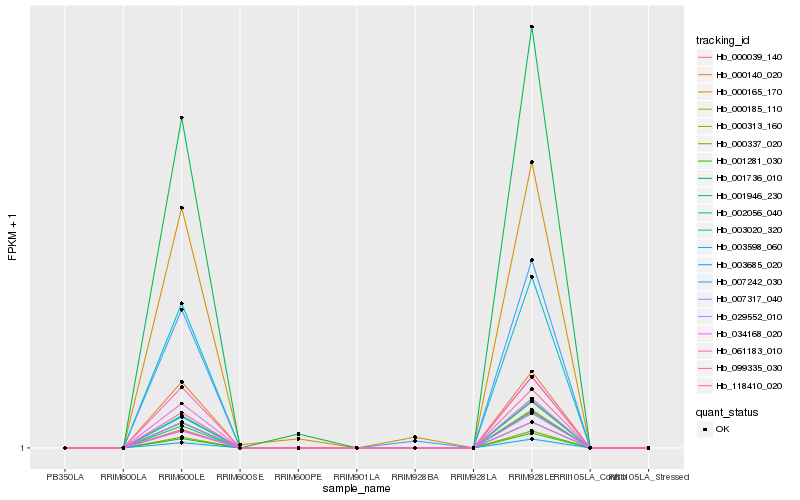
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007242_030 |
0.0 |
- |
- |
PREDICTED: methylesterase 3-like isoform X2 [Tarenaya hassleriana] |
| 2 |
Hb_000165_170 |
0.0827907735 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 3 |
Hb_001946_230 |
0.0843233614 |
- |
- |
- |
| 4 |
Hb_000313_160 |
0.0843288571 |
- |
- |
BnaC02g20570D [Brassica napus] |
| 5 |
Hb_000337_020 |
0.0846677005 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 6 |
Hb_061183_010 |
0.0847350339 |
- |
- |
PREDICTED: uncharacterized protein LOC105775353 [Gossypium raimondii] |
| 7 |
Hb_001281_030 |
0.0848578743 |
- |
- |
hypothetical protein POPTR_0010s18450g [Populus trichocarpa] |
| 8 |
Hb_003685_020 |
0.0862313871 |
- |
- |
PREDICTED: laccase-14-like [Fragaria vesca subsp. vesca] |
| 9 |
Hb_034168_020 |
0.0868993525 |
- |
- |
putative uncharacterized protein (mitochondrion) [Hevea brasiliensis] |
| 10 |
Hb_118410_020 |
0.0882393961 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C4 [Vitis vinifera] |
| 11 |
Hb_003020_320 |
0.0940850556 |
- |
- |
- |
| 12 |
Hb_000039_140 |
0.0983093415 |
- |
- |
nuclease, putative [Ricinus communis] |
| 13 |
Hb_099335_030 |
0.1000173175 |
- |
- |
hypothetical protein M569_10107, partial [Genlisea aurea] |
| 14 |
Hb_007317_040 |
0.1008431932 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Jatropha curcas] |
| 15 |
Hb_000140_020 |
0.101020114 |
- |
- |
BnaA02g09930D [Brassica napus] |
| 16 |
Hb_002056_040 |
0.1012775452 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 17 |
Hb_001736_010 |
0.1036200251 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_000185_110 |
0.1048005599 |
- |
- |
hypothetical protein JCGZ_11437 [Jatropha curcas] |
| 19 |
Hb_029552_010 |
0.106439155 |
- |
- |
hypothetical protein JCGZ_08684 [Jatropha curcas] |
| 20 |
Hb_003598_060 |
0.1070454331 |
- |
- |
hypothetical protein POPTR_0006s25370g [Populus trichocarpa] |