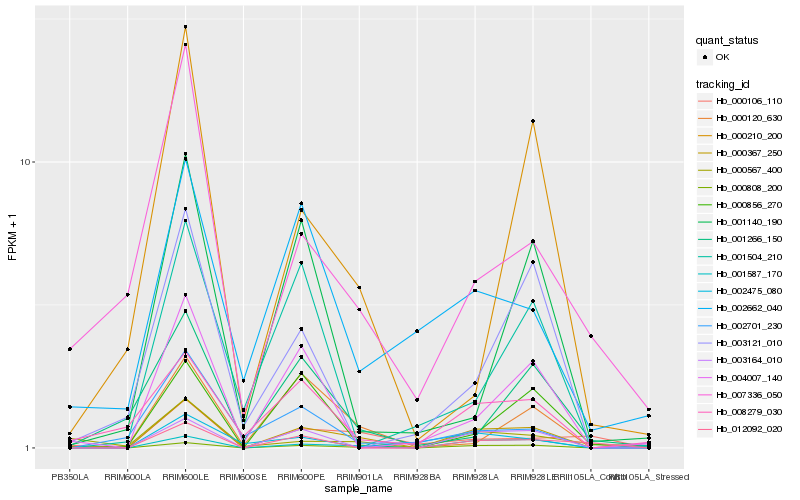
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_250 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_001587_170 |
0.1397570175 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 3 |
Hb_003164_010 |
0.2227573301 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000120_630 |
0.2697335404 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_000856_270 |
0.2804406391 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 6 |
Hb_004007_140 |
0.2869577464 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_008279_030 |
0.2896001042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012092_020 |
0.2923300764 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000210_200 |
0.2951685731 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 10 |
Hb_000808_200 |
0.3054631382 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1-like [Jatropha curcas] |
| 11 |
Hb_002701_230 |
0.3081447407 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |
| 12 |
Hb_003121_010 |
0.3107548167 |
- |
- |
hypothetical protein POPTR_0018s09720g [Populus trichocarpa] |
| 13 |
Hb_002475_080 |
0.3118622403 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 14 |
Hb_000567_400 |
0.3125605001 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 15 |
Hb_000106_110 |
0.3135684949 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 16 |
Hb_001140_190 |
0.3145744809 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 17 |
Hb_001266_150 |
0.3147452738 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 18 |
Hb_002662_040 |
0.3147477618 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 19 |
Hb_007336_050 |
0.3194268772 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 20 |
Hb_001504_210 |
0.3211087722 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |